Figure 4.
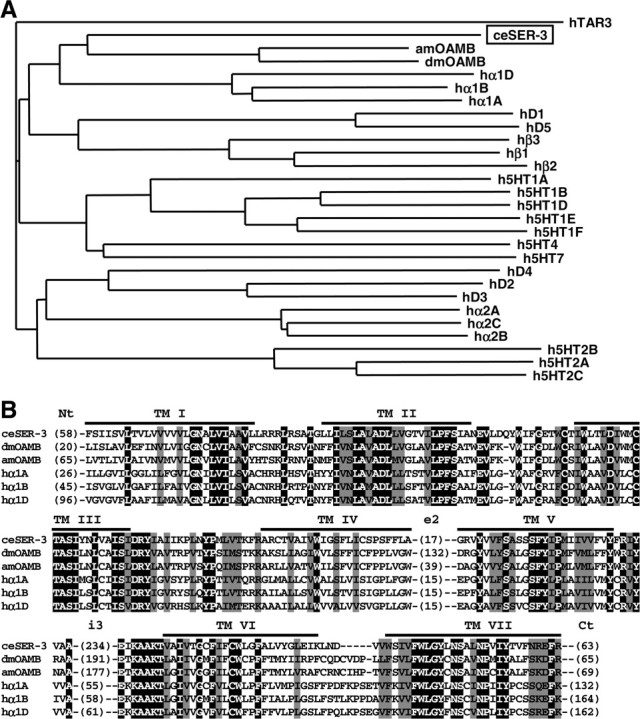
A phylogenetic tree and an alignment of invertebrate and human amine receptors. A, The deduced amino acid sequence of SER-3 was compared with invertebrate and human amine receptors using ClustalW, and a phylogenetic tree was calculated by the NJ method at DNA Data Bank of Japan web site. Relatively well conserved blocks (the region excluding the N terminus, second extracellular loops, third intracellular loops, and the C terminus) of sequences were used for the analysis. Receptor sequences used and the GenBank accession numbers are as follows: C. elegans SER-3 (ceSER-3, NP491954), Drosophila octopamine receptor (dmOAMB, AAC17442), Apis mellifera octopamine receptor (amOAMB, CAD67999), human dopamine receptors (hD1, P21728; hD2, P14416; hD3, P35462; hD4, P21917; and hD5, P21918), human serotonin receptors (h5HT1a, I38209; h5HT1b, JN0268; h5HT1d, A53279; h5HT1e, A45260; h5HT1f, A47321; h5HT2a, A43956; h5HT2b, S43687; h5HT2c, JS0616; h5HT4, Q13639; and h5HT7, A48881), and human adrenergic receptors (hα1A, NP000671; hα1B, NP000670; hα1D, NP000669; hα2A, A34169; hα2B, A37223; hα2C, A31237; hβ1, QRHUB1; hβ2, QRHUB2; and hβ3, QRHUB3). A human trace amine receptor 3 (hTAR3, AAO24660) was used as an out group. B, Alignment of SER-3 with insect OAMBs and human α1 adrenergic receptors. Amino acid sequences of C. elegans (ce) SER-3, Drosophila melanogaster (dm), and Apis mellifera (am) octopamine receptor OAMB and human α1 adrenergic receptors (hα1A, hα1B, and hα1D) were aligned with ClustalW. Residues that were identical and similar in all proteins are highlighted with black and gray, respectively. The positions of the putative transmembrane domains (TM) are indicated. The numbers in parentheses indicate the numbers of residues in the N terminus (Nt), second extracellular loops (e2), third intracellular loops (i3), and the C terminus (Ct), which were not aligned. The identities with ceSER-3 in transmembrane domains and flanking regions were as follows: dmOAMB, 51%; amOAMB, 53%; hα1A, 48%; hα1B, 45%; and hα1D, 41%.
