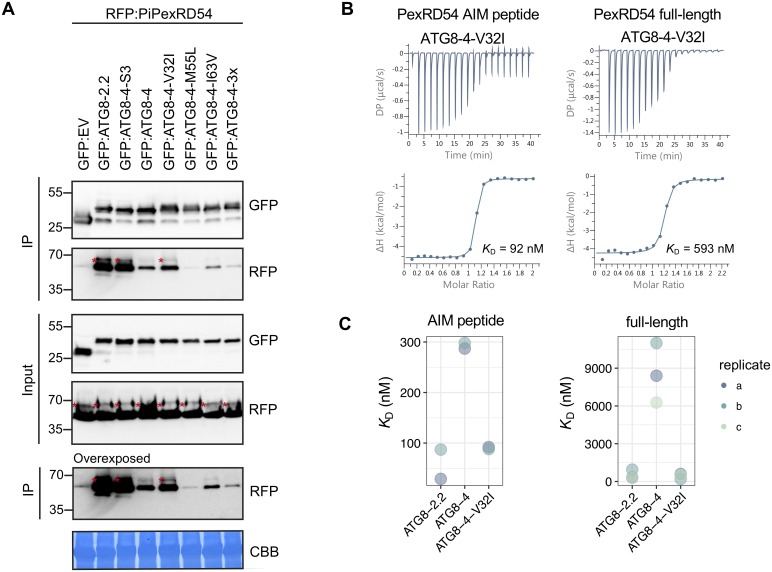
Fig 5. A single amino acid residue, Val-32 in the first β-strand, determines differential binding affinity of ATG8-4 towards PexRD54.
(A) Co-IP experiment between PexRD54 and all ATG8-4 point mutants. RFP:PiPexRD54 was transiently co-expressed with the controls GFP:EV, GFP:ATG8-2.2, GFP:ATG8-4-S3, and GFP:ATG8-4 and all of the GFP:ATG8-4 point mutants. IPs were obtained with anti-GFP antibody, and total protein extracts were immunoblotted with appropriate antisera (listed on the right of each). Stars indicate expected band sizes. (B) The binding affinity of ATG8-4-V32I with PexRD54-AIM peptide and full-length PexRD54 was determined by ITC. The top panels show heat differences upon injection of ligands and lower panels show integrated heats of injection (•) and the best fit (solid line) to a single site binding model using MicroCal PEAQ-ITC analysis software. (C) Chart summarizing the KD values for each interaction, including two replicates with the PexRD54 AIM peptide and three replicates with full-length PexRD54. ATG8, autophagy-related protein 8; CBB, Coomassie brilliant blue; Co-IP, co-immunoprecipitation; GFP:EV, green fluorescent protein empty vector; IP, immunoprecipitate; ITC, isothermal titration calorimetry; PiPexRD54, P. infestans PexRD54.