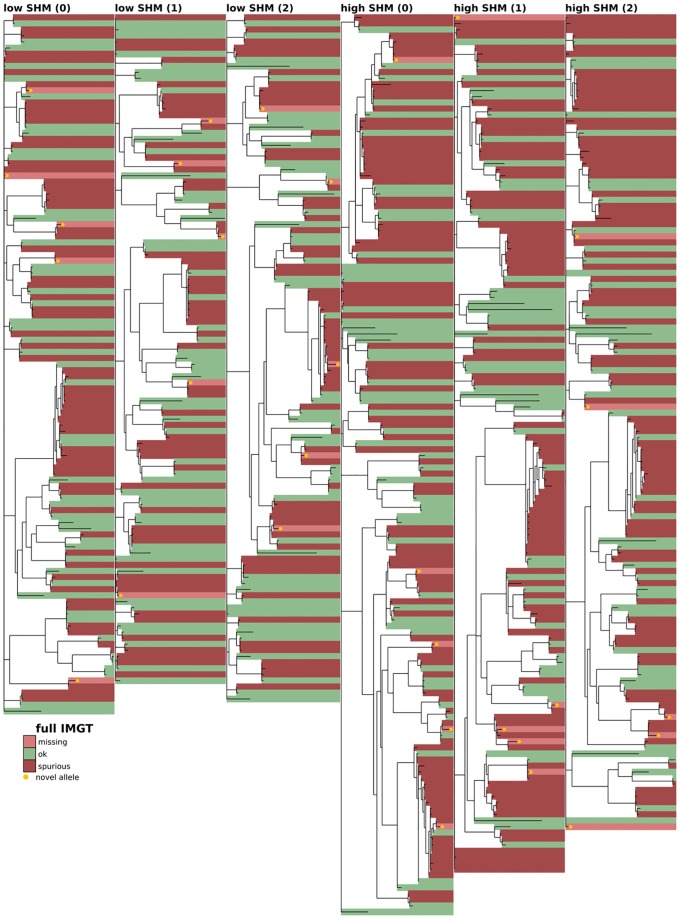
Fig 3. Full-repertoire germline set accuracy for the currently widespread method of aligning every sequence to its closest match in the full IMGT V gene set.
The phylogenetic tree is constructed with a leaf for each germline gene in either the true or inferred germline sets (see Methods). Branch lengths connecting different V gene families are set to zero. Leaves are colored according to the similarity of the true and inferred germline sets, with shared genes in green and unshared in red, the latter broken into missing (light red) and spurious (dark red). Novel alleles (not in the IMGT database, whether from the true simulated set or spuriously inferred) are highlighted in gold. Shown on the first three replicates (0-2) of both the low-SHM (left), and high-SHM (right) full-repertoire simulation samples (see text).