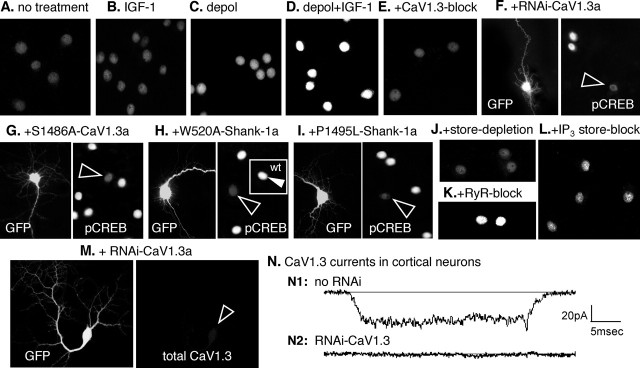
Figure 6.
CaV1.3a regulates pCREB in pyramidal neurons: requirements for IGF-1 potentiation of S1486, IP3 store release, and CaV1.3 interaction with Shank. A–L, Levels of activated pCREB were assayed by indirect immunofluorescence. To assess any regulation specifically attributable to CaV1.3 activity, all other calcium channel activity was inhibited pharmacologically (see Materials and Methods). To stimulate the activity of voltage-sensitive CaV1.3 channels, neurons were depolarized for 20 min in iso-osmotic 65 mm KCl, fixed immediately, and processed for immunocytochemistry (n = 8 independent experiments with each test condition assayed in triplicate). Changes in pCREB levels were analyzed using the Intensity module of the MetaMorph image analysis software; the fold change in pCREB was calculated using the mean level observed in the absence of any treatment (A) as the reference value of 1.0; values are means ± SEMs. A, In the absence of any treatment, pCREB levels were moderate/low. B, IGF-1 (20 ng/ml) alone has little effect. Fold change in pCREB levels were 1.04 ± 0.07. C, D, Depolarization in the absence of IGF-1 increases pCREB levels 1.6 ± 0.1-fold (C), but, in the presence of IGF-1, pCREB levels are very high (D), increasing by 3.1 ± 0.1-fold. E, F, Reducing CaV1.3 activity with 100 μm nimodipine (E) or CaV1.3a expression with RNAi specific for CaV1.3a (F; see Fig. 6M,N and supplemental Fig. 2 for controls) strongly reduces the elevation of pCREB induced by depolarization plus IGF-1. F, Left, The RNAi transfectant was identified by GFP coexpression. Right, pCREB levels are low in the transfectant (arrowhead) but high in two neighboring nontransfected neurons. Both treatments resulted in pCREB levels below that of the reference condition: high nimodipine, 0.8 ± 0.1; RNAi-CaV1.3a, 0.7 ± 0.1. G, Overexpressing S1486A-CaV1.3a prevents the IGF-1+ depolarization-induced increase in pCREB, producing a fold increase of only 1.3 ± 0.1. Left, Transfected neuron indicated by GFP fluorescence. Right, pCREB levels in all neurons in the same field with the GFP+ neuron (arrowhead). H, Requirement for Shank: overexpressing Shank-1a with a W520A point mutation to disrupt its SH3 domain blocks the ability of IGF-1+ depolarization to increase pCREB; the fold change in pCREB levels was 1.1 ± 0.1. Left, Transfected neuron (GFP). Right, pCREB levels in all neurons in the field with the W520A-Shank-1a+ neuron (open arrowhead). Inset, Unlike W520A-Shank-1a+ neurons, overexpressing wild-type (wt) Shank-1a in a sister culture results in high pCREB levels (filled arrowhead); the fold increase was 3.3 ± 0.1. I, Requirement for Shank–Homer interaction: overexpressing Shank-1a with a P1495L point mutation to disrupt its binding to Homer blocks the ability of IGF-1+ depolarization to increase pCREB; the fold change in pCREB was 0.9 ± 0.1. Left, Transfected neuron (GFP). Right, pCREB levels in all neurons in the field with the P1495L-Shank-1a+ neuron (open arrowhead). J–L, Ca2+ store dependence. Depleting stores with 1 μm thapsigargin (I) or blocking IP3-sensitive store release with 100 μm 2-APB (L) prevents the IGF-1+ depolarization-induced increase in pCREB, whereas inhibiting ryanodine (RyR)-sensitive store release (K) with 100 μm ruthenium red has little effect, producing a fold increase of 2.9 ± 0.1. For both store depletion and blocking IP3-sensitive store release, the fold change in pCREB levels was 0.9 ± 0.1. M, N, In cortical pyramidal neurons, RNAi to CaV1.3 strongly reduced CaV1.3 levels (M) and evoked CaV1.3 activity (N). M, Left, GFP cotransfection reveals a neuron expressing RNAi to CaV1.3a. Right, Total CaV1.3 (surface plus intracellular) in that neuron (open arrowhead indicates the soma). N, Somal CaV1.3 currents are evident in the absence of RNAi (N1) but undetectable in an RNAi-transfected neuron (N2). Recordings were performed in the presence of 1 μm nimodipine to inhibit CaV1.2 activity. See supplemental Fig. 2 (available at www.jneurosci.org as supplemental material) for additional information.