Abstract
Most forms of neuronal plasticity are associated with induction of the transcription factor Zif268 (Egr1/Krox24/NGF-IA). In a genome-wide scan, we obtained evidence for potential modulation of proteasome subunit and regulatory genes by Zif268 in neurons, a finding of significance considering emerging evidence that the proteasome modulates synaptic function. Bioinformatic analysis indicated that the candidate proteasome Zif268 target genes had a rich concentration of putative Zif268 binding sites immediately upstream of the transcriptional start sites. Regulation of the mRNAs encoding the psmb9 (Lmp2) and psme2 (PA28β) proteasome subunits, along with the proteasome-regulatory kinase serum/glucocorticoid-regulated kinase (SGK) and the proteasome-associated antigen peptide transporter subunit 1 (Tap1), was confirmed after transfection of a neuronal cell line with Zif268. Conversely, these mRNAs were upregulated in cerebral cortex tissue from Zif268 knock-out mice relative to controls, confirming that Zif268 suppresses their expression in the CNS. Transfected Zif268 reduced the activity of psmb9, SGK, and Tap1 promoter–reporter constructs. Altered psmb9, SGK, and Tap1 mRNA levels were also observed in an in vivo model of neuronal plasticity involving Zif268 induction: the effect of haloperidol administration on striatal gene expression. Consistent with these effects on proteasome gene expression, increased Zif268 expression suppressed proteasome activity, whereas Zif268 knock-out mice exhibited elevated cortical proteasome activity. Our findings reveal that Zif268 regulates the expression of proteasome and related genes in neuronal cells and provide new evidence that altered expression of proteasome activity after Zif268 induction may be a key component of long-lasting CNS plasticity.
Keywords: Egr-1, 26S proteasome, Lmp2, immunoproteasome, plasticity, proteasome activity
Introduction
Proteasomes are multi-subunit protein complexes that provide the major route of targeted protein degradation in cells. The subunit composition of the proteasome is thought to be constant across most cell types, although in immune cells, transcriptional induction of “inducible” subunit genes, the psmb8 (Lmp7), psmb9 (Lmp2), psmb10 (MECL-1), psme1 (PA28α), and psme2 (PA28β) genes, can change the subunit composition to form the “immunoproteasome” and hence increase proteasome activity (Harris et al., 2001; Cascio et al., 2002).
Although modifications in proteasome structure and function in the immune system have been studied intensively, little is known regarding proteasome adaptations to serve the highly specialized and polarized characteristics of neurons. However, the subunit composition of the proteasome varies in different brain regions (Noda et al., 2000; Ding and Keller, 2001). Interestingly, the pyramidal neurons of the cerebral cortex and hippocampus show the highest proteasome levels (Mengual et al., 1996) and express the inducible psmb9 and psmb8 subunits of the immunoproteasome constitutively (Diaz-Hernandez et al., 2003), linking psmb8 and psmb9 to the function of these glutamatergic neurons. Proteasome subunits are located in neuronal dendrites (Rezvani et al., 2003), implicating the proteasome in synaptic signaling.
The late phase of most, if not all, forms of synaptic plasticity is dependent not only on protein synthesis (Morris, 2004) but also on limited protein degradation. Inhibition of proteasome activity facilitates various forms of plasticity (Obin et al., 1999; Speese et al., 2003; Zhao et al., 2003; Juo and Kaplan, 2004). The proteasome regulates the levels of specific proteins with important roles in synaptic communication: NMDA receptor subunits, postsynaptic density scaffolding molecules (Colledge et al., 2003; Ehlers, 2003; Pak and Sheng, 2003), and synaptic glutamate transporters (Boehmer et al., 2003, 2004).
One of the few proteins with a confirmed role in many different forms of plasticity is the transcription factor (TF) Zif268. Zif268 is induced during periods of plasticity [i.e., during learning tasks or induction of long-term potentiation (LTP)] in the hippocampus, visual or temporal cortex, and spinal cord, in diverse species from primates to songbirds (Cole et al., 1989; Wisden et al., 1990; Jarvis et al., 1998; Kaczmarek et al., 1999; Mello et al., 2004; Morris, 2004). The threshold and duration of hippocampal LTP correlates with the degree of Zif268 induction (Richardson et al., 1992; Worley et al., 1993), and mice with targeted disruption of the Zif268 gene show deficits in late-phase LTP and in a range of memory tests (Jones et al., 2001), confirming the importance of Zif268 induction for long-term plasticity. In peripheral cells, Zif268 mediates transcriptional suppression or induction of various genes (Cao et al., 1993; Liu et al., 1996; Beckmann and Wilce, 1997; Thottassery et al., 1999; Bahouth et al., 2002; Virolle et al., 2003; Zhang and Liu, 2003; Wang et al., 2005). We have recently identified candidate target genes of Zif268 in neurons (James et al., 2005). A remarkable finding was that many of the genes identified encoded either components of the proteasome or proteins with functions closely allied to the proteasome. In this study, we test the hypothesis that the expression of proteasome subunits and functionally related genes is regulated by Zif268.
Materials and Methods
Expression constructs.
An overview of the plasmid constructs used during this study is provided in supplemental Table 1 (available at www.jneurosci.org as supplemental material). The Zif268 expression vector pZif268 has been described previously (Groot et al., 2000). As a control expression construct, an N-terminal deletion mutant (Δ3–372 aa) of pZif268, termed ptrZif268 (truncated Zif268), was constructed (James et al., 2005). This construct retains the 5′ and 3′ noncoding DNA sequence of the full-length Zif268 insert.
Promoter–reporter chloramphenicol acetyl transferase constructs.
Functional analysis of the serum/glucocorticoid-regulated kinase (SGK) (−1595 to +28), proteasome β9 subunit (psmb9) (−539 to +18), and Tap1 (−566 to −10) promoters [whereby +1 denotes the transcription start site (TSS)] was performed using plasmids containing the wild-type promoters cloned upstream of the chloramphenicol acetyl transferase (CAT) gene in pBLCAT3. The pTap1-CAT and pPsmb9-CAT constructs, whereby the rat Tap1/psmb9 intergenic region was subcloned in either orientation upstream of the CAT gene, were a gift from Dr. E. Blair (University of Leeds, Leeds, UK) (Proffitt and Blair, 1997). A region of the SGK promoter was PCR amplified from PC12 genomic DNA using primer pairs containing HindIII and XbaI sites, respectively, and directionally cloned into pBLCAT3 to give pSGK-CAT. Details of all vectors used are provided in supplemental Table 1 (available at www.jneurosci.org as supplemental material).
Animals and treatments.
The PC12 cell culture conditions were as described previously (James et al., 2004). Tissue corresponding to the entire striatum was isolated from male Wistar rats (200–300 g) that had received 1 mg/kg haloperidol (Tocris, Ellisville, MO) or vehicle (1 ml/kg of 0.5% hydroxypropylmethyl polyethylene glycol carbopol934) by intraperitoneal injection; 6 h later, striatal tissue was dissected and RNA was extracted as described below. Mice with targeted deletion of the Zif268 gene were a generous gift from Prof. P. Charnay (Institut National de la Santé et de la Recherche Médicale, Paris, France) (Topilko et al., 1998). We did not assess their behavior, but all animals were in the same behavioral conditions before they were killed. The anterior portion of the cerebral cortex (frontal cortex) incorporating prefrontal, cingulate, and motor areas was dissected for analysis.
Transient transfections.
Transient transfection of confluent PC12 cells has been described previously (James et al., 2004, 2005). Endotoxin-free DNA (2 μg of pZif268 or ptrZif268) was combined with Lipofectamine 2000 (LF200; Invitrogen, Paisley, UK) in OPTI-MEM I medium incubated at room temperature for 20 min. RNA was extracted 48 h after transfection.
GeneChip data analysis.
3 GeneChips were used for each group. The average difference and expression level of genes were calculated according to absolute and comparison analysis algorithms according to Affymetrix (Santa Clara, CA) protocols. Normalization was performed using BioConductor (http://biowww.dfci.harvard.edu/∼bioconductor/). Significant changes in expression levels for the pZif268 transfection treatment relative to the control, ptrZif268, treatment were calculated using Significance Analysis of Microarrays (SAM) (Tusher et al., 2001) (www-stat.stanford.edu/∼tibs/SAM/). SAM uses permutations of the repeated measurements to provide an accurate estimate of the percentage of genes identified by chance, the false discovery rate (FDR), and hence avoids the usual problems of a large number of false positive results because of the performance of multiple uncorrected statistical tests. The FDR was set at a threshold of 0.05 in our analysis, so few false positive results are anticipated.
In silico inspection of promoter regions of candidate target genes.
The program Gene2Promoter (Genomatix Software, Munich, Germany) was used to extract the promoter regions from a list of accession numbers. Typically, a region of 1101 bp (−1000 to +100) was extracted. Zif268 binds to consensus DNA sequences known as Egr response elements (EREs). The program MatInspector (Quandt et al., 1995) was used to locate putative EREs in a DNA sequence. Core similarity was set at the default level of 0.75. A selection of randomly chosen genes was obtained using the random gene selection tool at the Regulatory Sequence Analysis Tools web site provided by the Service de Conformation des Macromolécules Biologiques et de Bioinformatique (l’Université Libre de Bruxelles, Bruxelles, Belgium).
Reverse transcription-PCR.
Where possible, at least one of the primer pairs used for PCR was designed to span an exon–intron boundary. Primer sequences are provided in Table 1. RNA was extracted from PC12 cells, mouse frontal cortex, or rat striatum using the RNeasy Mini kit (Qiagen, Chatsworth, CA). A single PCR cycle within the logarithmic phase of the PCR was selected. Band intensity was normalized to the corresponding signal for the “housekeeping” gene glyceraldehyde-3-phosphate dehydrogenase (Gapdh), and differences between the groups were assessed using the two-sample t test (striatal/cortical studies) and the Mann–Whitney U test (PC12 validation study).
Table 1.
RT-PCR primer sequences used during this study
| Gene targeta | Oligonucleotidesb
|
Gene/locus accession numberc | Amplicon size (bp) | |||
|---|---|---|---|---|---|---|
| Sense |
Antisense |
|||||
| Exons | Sequence 5′–3′ | Exons | Sequence 5′–3′ | |||
| Gapdh | 1 | CCGCTGATGCCCCCATGTTTGTGAT | 1 | GGCATGTCAGATCCACAACGGATAC | 24383 | 369 |
| psme2 | 6–7 | AAGTGCATTCTGGTAATCACATGGATC | 10–11 | GATGAAGCTCAGCATAGAAGGCTCTC | 29614 | 304 |
| Mouse psme2 | 6–7 | AAGTGCATTCTGGTAATCACATGGATC | 10–11 | GATAAAGCTCAGCATAGAAGGCCCTC | 19188 | 304 |
| psmb9 | 1–2 | AGTCCACACCGGGACAACCATCATG | 5–6 | CCAGAGTGATGGCGTCTGTGGTGAA | 24967 | 497 |
| SGK | 3–4 | AACCCCTCACCTCCTCCAAGTCCCTCTCA | 8–9 | AGGAGCGAGATACTCAGGCGTGCCACA | 29517 | 585 |
| Tap1 | 5–6 | TGGACCATGAGTGTCTCGGGAATGC | 9–10 | CAGTTTCACCTACCTCTGTGTCATAGCCCT | 24811 | 682 |
aPrimers target rat cDNA template unless otherwise denoted.
bWhere possible, at least one of the primer pairs were predicted to span an exon–intron boundary. The exon number for which a section of the primer sequence is predicted to target are denoted. For primers predicted to span exon–intron boundaries, the portions of the primer sequence that target distinct exons are characterized by alternating type style (roman or italic/underline).
cThe Entrez Gene ID is provided. The exon–intron structure information and sequence template for primer design were also obtained from the ENSEMBL database (www.ensembl.org).
Reporter assays.
Transfections with either pZif268 or ptrZif268 expression plasmids were performed as described above, along with a reporter plasmid (pPsmb9-CAT, pSGK-CAT, or pTap1-CAT; 2 μg per well) and the internal reference plasmid, driving β-galactosidase expression from a cytomegalovirus (CMV) promoter, pCMVβ (0.5 μg per well) (see supplemental Table 1, available at www.jneurosci.org as supplemental material). Reporter expression was assayed 48 h after transfection. β-Galactosidase activities were also determined and used to correct for variations in transfection efficiencies. Differences between groups were assessed using one-way ANOVA, and differences relative to control values were assessed using 95% confidence intervals of the mean and the Wilcoxon signed rank test.
Western blotting.
Polyvinylidene difluoride membranes (Invitrogen) were blocked and probed overnight at 4°C with anti-psmb9 (1:500 dilution; Abcam, Cambridge, UK). After incubation in peroxidase-conjugated goat anti-rabbit IgG (Jackson ImmunoResearch, West Grove, PA), diluted to 0.08 pg/ml, and further washing, immunoreactive bands were revealed using ECL Plus on Hyperfilm ECL (Amersham Biosciences, Little Chalfont, UK). Integrative optical density readings were obtained using NIH Image (W. Rasband, National Institutes of Health, Bethesda, MD) as described previously (Morris, 1995; Simpson and Morris, 2000; Conway et al., 2004), and differences relative to control values were assessed using the Wilcoxon signed rank test.
Proteasome assay.
Protein concentrations were determined using the Bio-Rad (Hercules, CA) microprotein assay system and normalized. 20S proteasomal activity was quantified by monitoring the accumulation of the fluorescent cleavage product 7-amino-4-methylcoumarin (AMC) from the synthetic proteasomal substrate Suc-Leu-Leu-Val-Tyr-aminomethylcoumarin (LLVY-AMC; Calbiochem, La Jolla, CA). Each whole-cell lysate (PC12 cells) or tissue extract (mouse cortex) was preincubated for 2 min at 37°C with 40 μm LLVY-AMC before quantification of the fluorescent proteolysis product in a Fluoroskan Ascent microplate fluorometer (Thermo Labsystems, Basingstoke, UK). The fluorescence of the released AMC was measured (every 30 s for 30 min) using an excitation wavelength of 340 nm and an emission wavelength of 450 nm. The concentration of liberated products was calculated using a standard curve for AMC and significance assessed by three-way ANOVA (factors: treatment, culture and time).
Results
Microarray analysis of genes encoding proteasome subunits and functionally related genes after overexpression of Zif268 in PC12 cells
After increased expression of Zif268 in PC12 neurons, we assessed the changes in the pattern of gene expression using Affymetrix GeneChips to profile a large proportion of the neuronal transcriptome. In a previous report, we noted that a number of proteasome and related genes were prominent in the list of putative neuronal Zif268 target genes (James et al., 2005). The proteasome comprises a 20S core consisting of 28 subunits composed of structural proteasome (psm) α subunits (psma1–7) and proteolytic β subunits (psmb1–7), with a 19S regulator at each end consisting of ATPase subunits (psmc1–6) and non-ATPase subunits (psmd1–14). In the immune system, interferon-mediated transcriptional induction of the psme1 (PA28α) and psme2 (PA28β) genes replaces the 19S regulator at one or both ends with the 11S or PA28 regulator (Cascio et al., 2002). Immune challenge also increases transcription of the inducible β subunits psmb8, psmb9, and psmb10, which then replace the constitutive β subunits psmb5, 6, and 7. The presence of the 11S regulator and the psmb8 and psmb9 subunits in the immunoproteasome then increases proteasome activity (Harris et al., 2001; Cascio et al., 2002). Detailed analysis revealed that increased expression of Zif268 suppressed the expression of a number of proteasome subunit genes, including psmc4, psmb8, psmb9, psme1, and psme2 (Fig. 1a).
Figure 1.
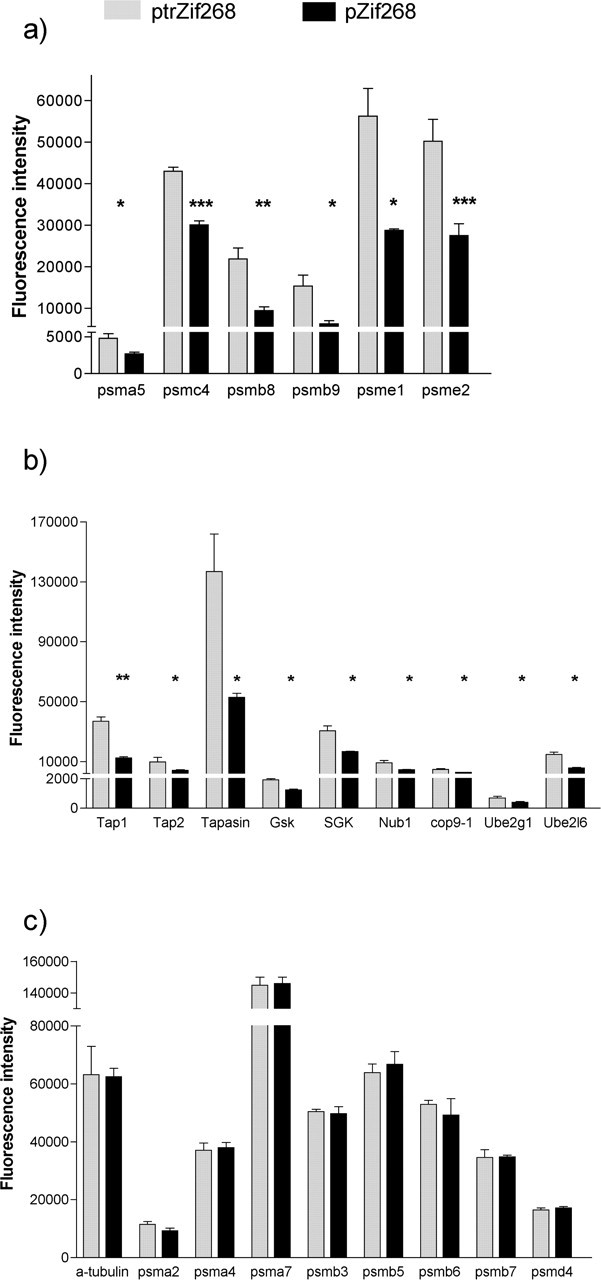
Microarray analyses of proteasome gene expression in Zif268-transfected versus control-transfected PC12 cells. Affymetrix microarray analysis of proteasome and related genes in PC12 cells after transfection with the Zif268 expression vector pZif268 (▪) or the inactive mutated Zif268 expression vector ptrZif268 (▦) is shown. a, Proteasome subunit genes showing significant differential expression. b, Proteasome-related genes showing significant differential expression. c, Proteasome subunit genes that did not show significant differential expression, with α-tubulin for comparison. Results are mean ± SEM. *p < 0.05, **p < 0.01, and ***p < 0.001 versus inactive Zif268 transfection (SAM analysis; n = 3).
In addition, expression of a number of proteasome accessory and regulatory genes was also decreased, including proteasome-related kinases such as SGK, E2 and E3 ubiquitin ligases, which are key intermediates in the tagging of proteins for proteasomal degradation, and genes encoding components of the COP9 signalosome (Fig. 1b). The levels of the mRNAs derived from the three genes encoding subunits of the proteasome-associated antigen peptide transporter (Tap) complex, Tap1, Tap2, and tapasin, also appeared decreased. However, the level of expression of other proteasome subunit genes was not affected by the increased expression of Zif268 (Fig. 1c).
Bioinformatic analysis of Zif268-regulated proteasome genes
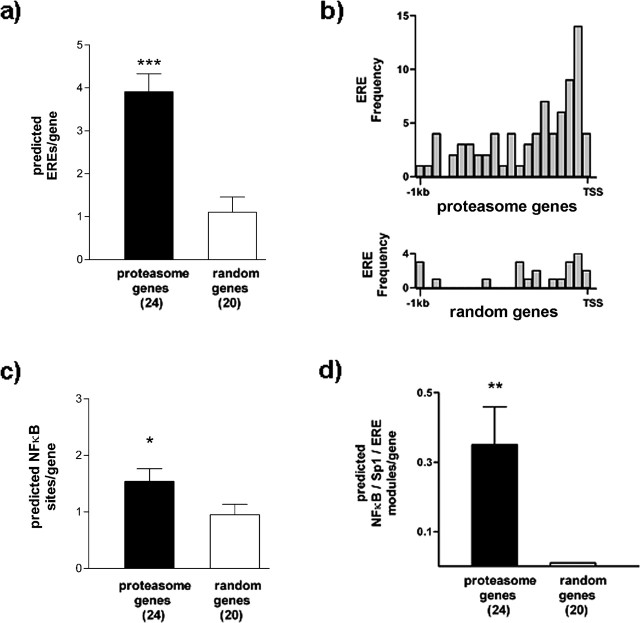
Bioinformatic analysis of gene promoters, using software trained to identify consensus TF recognition sequences, has proved of great use in detecting functional TF binding sites (Tullai et al., 2004; Chowdhary et al., 2005; Kamalakaran et al., 2005; Xie et al., 2005; Zhang et al., 2005). To assess the likelihood that the identified genes are direct targets of Zif268, we assessed the abundance of consensus EREs in the genomic region 1 kb upstream of the TSS of 24 proteasome-related genes identified as potential Zif268 targets compared with 20 randomly selected genes (see Materials and Methods) (Table 2), using the program MatInspector (Quandt et al., 1995). These analyses revealed, in all cases, consensus EREs in the promoter regions of the 24 proteasome-related genes. Indeed, in many cases, multiple potential EREs were detected (Fig. 2a, Table 2).
Table 2.
Putative Zif268-regulated proteasome genes and randomly selected genes used for the bioinformatic analysis
| Proteasome genes | ERE | Sp1 | NFκB | Randomly selected genes | ERE | Sp1 | NFκB |
|---|---|---|---|---|---|---|---|
| anapc1 | 2 | 2 | 0 | NM_001916.2 | 0 | 1 | 1 |
| cdc5-like | 2 | 5 | 3 | BRD4 | 0 | 4 | 2 |
| cop1 | 6 | 5 | 1 | C14orf15 | 0 | 3 | 2 |
| Herc3 | 3 | 2 | 2 | C20orf26 | 0 | 11 | 0 |
| mdm2 | 4 | 5 | 1 | DKFZp761G058 | 1 | 3 | 2 |
| Nedd4 | 2 | 5 | 1 | FLJ20343 | 2 | 1 | 1 |
| Nedd8-UB1 | 8 | 7 | 4 | HEG | 0 | 4 | 2 |
| psma5 | 1 | 8 | 1 | KIAA1958 | 4 | 5 | 0 |
| psmb8 | 5 | 4 | 1 | LIN7C | 1 | 2 | 1 |
| psmb9/Tap1 | 4 | 6 | 3 | LOC148003 | 0 | 1 | 1 |
| psmc4 | 2 | 13 | 2 | LOC388524 | 0 | 2 | 1 |
| psme1 | 3 | 1 | 0 | LOC388617 | 0 | 18 | 0 |
| psme2 | 8 | 14 | 2 | LOC392539 | 0 | 14 | 0 |
| Rhysin2 | 3 | 2 | 2 | LOC401352 | 2 | 3 | 2 |
| SAK | 3 | 0 | 0 | LOC402055 | 1 | 27 | 0 |
| SGK | 7 | 9 | 0 | MAC30 | 2 | 2 | 2 |
| sgt | 3 | 1 | 1 | PCDHA10 | 0 | 11 | 0 |
| Tap2 | 3 | 10 | 3 | RCN2 | 6 | 1 | 1 |
| tapasin | 7 | 11 | 3 | ZNF26 | 2 | 1 | 0 |
| ubc7 | 5 | 1 | 1 | ZNF530 | 1 | 1 | 1 |
| UBE2l6 | 2 | 6 | 2 | ||||
| ubiquilin | 5 | 4 | 2 | ||||
| ubiquitinD | 2 | 2 | 1 | ||||
| vhl | 4 | 3 | 1 |
The candidate Zif268-regulated proteasome genes were compared with a randomly selected set of genes (see Materials and Methods) for the number of potential EREs, Sp1 sites, and NFκB sites present in the upstream regions. For details of procedures, see Materials and Methods. The consensus sequences are as follows: ERE, TGCGTRGGCGK; Sp1, NGGGGGCGGGGYN; NFκB, SGGRNWTTCC. Note that bioinformatic predictions are not in themselves indicative of functional significance.
Figure 2.
Bioinformatic analysis of candidate Zif268-regulated proteasome genes. The 1 kb upstream regions from the putative Zif268-regulated proteasome genes were compared with the equivalent regions from 20 randomly selected genes (see Materials and Methods and supplemental Table 2, available at www.jneurosci.org as supplemental material). The numbers of genes are shown in parentheses. a, The number of predicted EREs is significantly greater in the proteasome genes. ***p < 0.001 versus randomly selected genes (Mann–Whitney U test). b, Frequency distribution of EREs in proteasome genes (top) and randomly selected genes (bottom). EREs are concentrated immediately upstream of the TSS in the proteasome genes. c, The number of predicted NFκB binding sites is significantly greater in the proteasome genes. *p < 0.05 versus randomly selected genes (Mann–Whitney U test). d, The number of predicted NFκB/Sp1/ERE modules is significantly greater in the proteasome genes. For the randomly selected genes, no NFκB/Sp1/ERE modules were detected. **p < 0.001 versus 0 (Wilcoxon test). Results are shown as mean ± SEM.
This is compatible with evidence that functional Zif268/Egr1 sites are generally located in proximal promoter regions. Although the presence of a consensus binding site in the promoter region does not of itself prove that the gene is a target of the TF, this is nonetheless consistent with these genes being direct targets of Zif268. Considering that EREs are underrepresented in promoters across the entire genome (Kel-Margoulis et al., 2003; Xie et al., 2005), there is a remarkable concentration of potential EREs in the promoter regions of these genes.
The predicted ERE abundance was substantially greater in the 1 kb upstream regions of the proteasome genes relative to the randomly selected genes, suggesting that this finding did not occur by chance (Fig. 2a). To complement this result, we also used another approach to assess the representation of EREs in the promoter regions of the proteasome-related genes identified as Zif268 targets. Rather than looking for specified consensus TF recognition sites in individual promoter sequences, the “overrepresented TF binding site prediction” method (Zheng et al., 2003) searches, with no a prioiri assumptions, for significantly overrepresented cis elements in a group of sequences and performs post hoc matches to the TRANSFAC database. This method identified an enrichment of Sp1 sites [P(M,t) = 2.8 × 10−10] and EREs [P(M,t) = 3.5 × 10−8] in the promoter regions of the proteasome-related genes. No enrichment of Sp1 sites or EREs was detected in the randomly selected promoter sequences.
We also assessed the distribution of the EREs in these putative promoter regions, because, in many known Zif268 target genes, EREs are located close to the TSS. We found a notable concentration of the putative EREs immediately upstream of the TSS from the proteasome genes (Fig. 2b). Because some proteasome genes are thought to be regulated at the transcriptional level by members of the nuclear factor κB (NFκB)/Rel family of TFs (Chatterjee-Kishore et al., 1998; Whitehouse and Tisdale, 2003; Marques et al., 2004), and because Zif268 is known to interact with and modulate NFκB/Rel transactivation ability (Cogswell et al., 1997; Chapman and Perkins, 2000), we similarly monitored the abundance of predicted NFκB/Rel binding sites in the 1 kb upstream regions of the proteasome-related genes. These sites were also enriched in the upstream regions of the proteasome genes relative to the randomly selected genes (Fig. 2c, Table 2), although, interestingly, the relative abundance of predicted EREs was even greater than that of the NFκB/Rel binding sites (Fig. 2a,c).
The TF Sp1 has also been implicated in the regulation of transcription of proteasome genes, and there is considerable evidence to suggest that many actions of Zif268 involve competition or interaction with Sp1 (Ebert et al., 1994; Srivastava et al., 1998; Fukada and Tonks, 2001; Davis et al., 2003; Tan et al., 2003). We therefore assessed the association between predicted EREs and Sp1 sites in the upstream regions of the proteasome genes. We found that 20 of 24 proteasome-related genes (83%) contained overlapping EREs and Sp1 sites in these upstream regions compared with 2 from 20 randomly selected genes (10%). The relationship between the Sp1 sites and EREs for four prototypical genes is shown in supplemental Fig. 1 (available at www.jneurosci.org as supplemental material). To test the hypothesis that the ability of Zif268 to alter the expression of these genes might involve coordinate interaction between Sp1, NFκB/Rel, and Zif268, we searched for possible modules of TF binding sites containing these three motifs in close proximity (<30 bp). We found that 8 from 24 proteasome genes showed the presence of one or more copies of this module in their upstream region compared with none of the randomly selected genes (Fig. 2d). It is thought that NFκB/Rel proteins and Zif268 can interact over larger distances (Wright et al., 1995). Although we did not analyze more widely separated Sp1, NFκB/Rel, and Zif268 modules in detail, it was clear that a less conservative criterion (>30 bp) would have revealed the presence of this combination of motifs in many more proteaseome gene upstream regions.
These results suggest that Zif268 may exert a previously unsuspected regulatory control on proteasome gene expression in neurons and could interact in some cases with the relatively well characterized NFκB/Rel and Sp1 regulatory pathways. This control appears to be exerted at multiple levels of proteasome function: on genes encoding components of the proteasome, proteins mediating peptide export to the class I MHC, ubiquitin ligase enzymes, and proteins involved in proteasome regulation.
Validation of the influence of Zif268 on expression of proteasome subunit genes and proteasome-related genes
Thus far the data clearly implicated Zif268 in the coordinate regulation of a number of proteasome subunit and proteasome-related genes. Potential regulation of proteasome subunit genes and associated E2 and E3 ligases is striking in light of the emerging concept that altered proteasome function affects synaptic plasticity. The altered expression of inducible proteasome subunits such as psmb9 and psme2, which are known to be transcriptionally regulated in the immune system to alter the function of the proteasome, is especially prominent. Potential regulation of SGK by Zif268 is also of particular interest, because SGK protects synaptic glutamate transporters from proteasomal degradation (Boehmer et al., 2003, 2004; Schniepp et al., 2004) and altered glutamate transporter levels have been suggested to contribute to LTP and memory (Levenson et al., 2002; Shen and Linden, 2005). Similarly, the oligopeptides generated by proteasomal protein digestion are transported into the endoplasmic reticulum via the antigen transporter complex (Tap1, Tap2 and tapasin). The TAP complex is physically associated with the proteasome in many cell types (Hwang et al., 2001), and, intriguingly, targeted deletion of the Tap1 gene in mice contributes to deficits in synaptic plasticity (Huh et al., 2000).
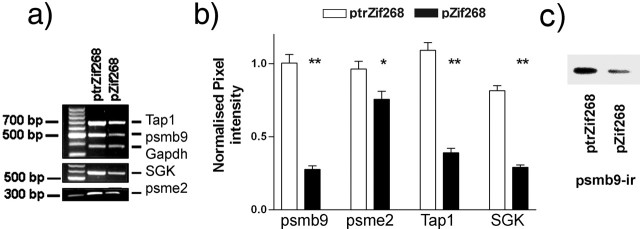
Four genes involved in different stages of proteasome function were selected for additional investigation: psmb9 and psme2, which encode proteasome subunits known to modulate proteasome activity; Tap1, which is physically associated with the proteasome and has been implicated in synaptic plasticity; and SGK, which regulates the access to the proteasome of certain proteins involved in plasticity and is also itself degraded by the proteasome. We validated the differential expression of these four candidate Zif268 target genes (psmb9, psme2, SGK, and Tap1) after Zif268 transfection of PC12 cells using reverse transcription-PCR (RT-PCR) (Fig. 3a,b). The levels of all the mRNAs tested were suppressed by Zif268 transfection, consistent with the results from the microarray experiment. We also monitored the levels of psmb9 protein immunoreactivity by Western blotting. Although there was greater variability in the levels of protein compared with the mRNA levels, the results suggested that the levels of psmb9 protein were also regulated by Zif268 transfection (Fig. 3c).
Figure 3.
Validation of psmb9, psme2, Tap1, and SGK regulation by Zif268 in PC12 cells. Semiquantitative RT-PCR was used to determine the relative expression levels of the genes. a, Representative RT-PCR product band intensities in pZif268- and ptrZif268-transfected PC12 cells. Note the stronger band intensities for Tap1, psmb9, SGK, and psme2 in the cells transfected with the truncated Zif268 expression vector ptrZif268. b, Relative amounts of target cDNA were determined (see Materials and Methods) 48 h after transfection. Results from five independent experiments are shown from ptrZif268 (□) and pZif268 (▪)-transfected cells (means ± SEM). *p ≤ 0.05; **p < 0.01, expression levels lower in pZif268-transfected PC12 cells compared with ptrZif268-transfected cells (Mann–Whitney U test). c, Levels of psmb9 protein immunoreactivity (psmb9-ir) in pZif268- and ptrZif268-transfected PC12 cells. The results suggested a corresponding regulation of protein levels.
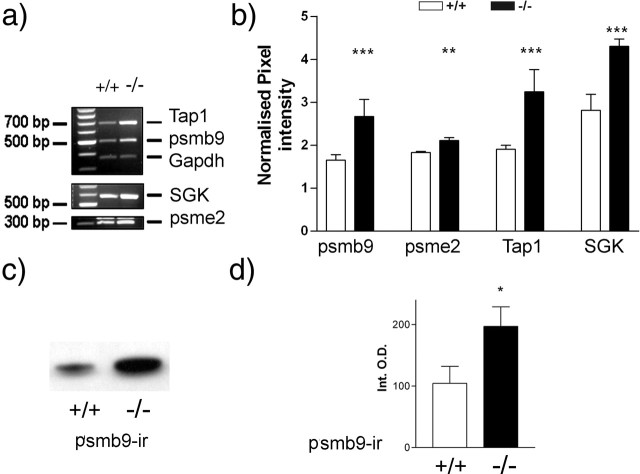
The level of expression of these genes was also tested in cerebral cortex tissue from mice with a targeted deletion of the Zif268 gene (Zif268 knock-out mice) and wild-type control mice. The levels of psmb9, psme2, SGK, and Tap1 mRNAs were all found to be increased in the tissue from the Zif268 knock-out mice (Fig. 4a,b). In the case of psmb9, we also tested whether this regulation of expression at the mRNA level was mirrored by altered expression at the protein level. In cortical tissue from the Zif268 knock-out mice, there was an increase in psmb9 protein levels relative to wild-type mice (Fig. 4c,d). We also obtained evidence for elevated SGK expression in the hippocampus from Zif268 knock-out mice relative to controls (data not shown).
Figure 4.
Validation of psmb9, psme2, Tap1, and SGK regulation by Zif268 in Zif268 knock-out mice. Semiquantitative RT-PCR was used to determine the relative expression levels of the genes. a, Representative RT-PCR product band intensities for tissue from wild-type (+/+) and Zif268 knock-out (−/−) mice. b, Relative mRNA expression in cortical tissue isolated from age-matched Zif268 wild-type (□) and Zif268 knock-out (▪) mice. c, Western blot bands showing psmb9 immunoreactivity in cortical tissue from wild-type (+/+) and Zif268 knock-out (−/−) mice. d, Relative amounts of psmb9 immunoreactivity in cortical tissue isolated from age-matched Zif268 wild-type (□) and Zif268 knock-out (▪) mice. The results are normalized pixel intensity (relative to Gapdh signal intensity) (b) or integrated band optical density (d) and are mean ± SEM: *p < 0.05, **p < 0.01, ***p < 0.001, expression levels greater in knock-out mice compared with wild-type mice (Mann–Whitney U test).
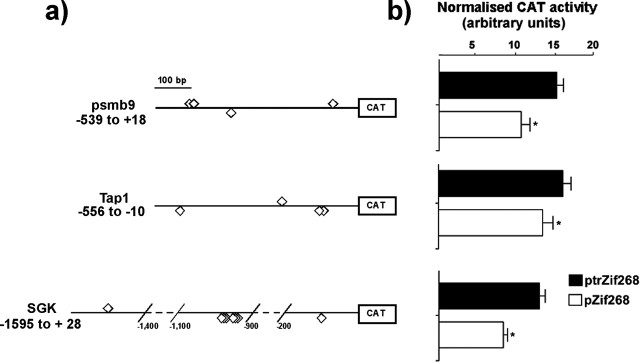
Assessment of ability of Zif268 to transactivate the SGK, Tap1, and psmb9 promoters
Althougb the psme2 promoter is not well characterized, the structures of psmb9, SGK, and Tap1 promoters have been described previously (Webster et al., 1993; Proffitt and Blair, 1997; Waldegger et al., 1998). We therefore focused on these three genes, each of them with a potentially interesting role in synaptic plasticity. Promoter–reporter CAT constructs for the potential Zif268 target genes [all of which contain a putative ERE site(s) in their promoter regions] were prepared (Fig. 5a) to test the ability of Zif268 to alter the activity of the promoters. PC12 cells were cotransfected with these constructs and either full-length Zif268 or control, truncated Zif268. In the presence of full-length Zif268, promoter activity was suppressed for each of the three genes selected (Fig. 5b).
Figure 5.
Promoter activity of candidate Zif268 target genes. a, Plasmids used for the promoter activity study (see Materials and Methods). The positions of putative ERE sites are indicated (⋄). For simplicity, lengthy regions of the SGK promoter devoid of putative EREs are represented as dashed lines. b, Plasmids (2 μg) containing wild-type promoter regions of the psmb9, SGK, and Tap1 genes were cotransfected in PC12 cells with 2 μg of either pZif268 (expressing Zif268; □) or, as control, ptrZif268 (expressing truncated Zif268; ▪). In each experiment, values were corrected for transfection efficiency (see Materials and Methods). Results from at least five independent experiments are shown (means ± SEM). *p ≤ 0.05 versus control (Wilcoxon signed rank test).
Note that although the bioinformatic analysis supports the possibility of direct actions in each case, it should be remembered that there is a possibility that the effects of Zif268 could be indirect, reflecting the induction of other TFs by Zif268, which then themselves act on the SGK, Tap1, and psmb9 promoters. However, whether the effects of Zif268 are direct or indirect, our results clearly show that altered expression of proteasome and related genes is likely to be a consequence of the elevated Zif268 expression that occurs during neuronal plasticity.
Zif268-regulated proteasome genes and CNS plasticity
We extended our analysis of the regulated expression of these genes using a well characterized model of CNS plasticity, the response of rat striatal neurons to elevated corticostriatal activity after D2 receptor blockade in vivo. Systemic administration of drugs with D2 dopamine receptor antagonist properties, such as haloperidol, increases glutamate release from corticostriatal terminals and produces LTP of corticostriatal transmission (Calabresi et al., 1997; Cepeda et al., 2001), with dramatic induction of striatal Zif268 (with a peak expression of ∼1–2 h) (Nguyen et al., 1992; Simpson and Morris, 1994; Keefe and Gerfen, 1995). We confirmed induction of Zif268 after haloperidol administration (Fig. 6a). We also determined the relative expression levels of the SGK, psmb9, and Tap1 mRNAs 6 h after haloperidol administration to rats. All three genes were found to be differentially expressed in the striatum relative to control animals (Fig. 6b,c). Intriguingly, although psmb9 and Tap1 were downregulated in the striatal neurons, SGK expression was upregulated.
Figure 6.
Expression of Zif268-target candidate genes during in vivo neuronal plasticity. Semiquantitative RT-PCR was used to determine the relative expression levels of the psmb9, SGK, and Tap1 genes in striatal tissue isolated from haloperidol-treated rats (hal) compared with corresponding vehicle-treated controls (veh). a, Levels of striatal Zif268 mRNA 30 min after treatment with vehicle (□) or haloperidol (▪). b, Representative RT-PCR product band intensities for candidate genes expressed in striatum tissue for haloperidol- and vehicle-treated rats. c, Relative amounts of target cDNA were determined (see Materials and Methods) 6 h after treatment with vehicle (□) or haloperidol (▪). The results shown are mean ± SEM (n = 3–6). *p ≤ 0.05; **p ≤ 0.01 (two-sample t test).
Assessment of ability of Zif268 to modify proteasome activity
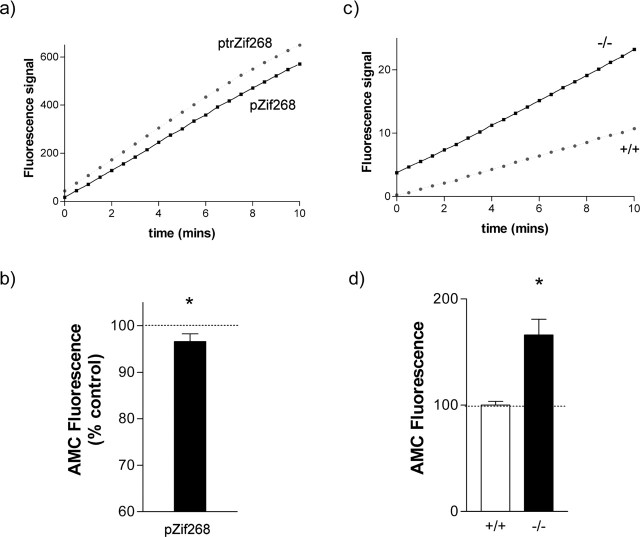
Proteasome activity in transfected PC12 cells was monitored using the fluorogenic synthetic proteasome substrate LLVY-AMC. Generation of fluorescent AMC in these studies was completely blocked by the proteasome inhibitor MG132 (N-carbobenzoxyl-Leu-Leu-leucinal) (10 μm) (data not shown). It was found that full-length Zif268 induced a small but clear decrease in the generation of AMC by the proteasome relative to truncated, inactive Zif268 (Fig. 7a,b).
Figure 7.
Effect of Zif268 on proteasome activity. a, Liberation of fluorescent AMC by the proteasome over time from a PC12 cell culture transfected with control ptrZif268 (black squares) or active Zif268 (gray circles). In five cultures, there was a highly significant effect of treatment as assessed by three-way ANOVA (factors: culture, treatment, and time; effect of treatment, F(1,544) = 75.83; p < 0.001). b, The amount of AMC liberated in ptrZif268- versus pZif268-transfected cells at 20 min, expressed as a percentage of the AMC signal in the control (ptrZif268-transfected) culture. *p < 0.05 versus control (Wilcoxon test). c, Liberation of fluorescent AMC by the proteasome over time from cerebral cortex tissue from a wild-type mouse (black squares) or a Zif268 knock-out mouse (gray circles). In four mice from each genotype, there was a highly significant effect of genotype as assessed by two-way ANOVA (factors: time and genotype; effect of genotype, F(1,426) = 24.80; p < 0.001). d, The amount of AMC liberated in cortical tissue from wild-type (+/+; □) and Zif268 knock-out (−/−; ▪) mice at 20 min, expressed as a percentage of the AMC signal in the control (wild-type) tissue. *p < 0.05 versus control (Wilcoxon test). Results are shown as mean ± SEM.
Proteasome activity was similarly monitored in cortical tissue from Zif268 knock-out mice (Topilko et al., 1998) and corresponding wild-type controls. Proteasome activity was substantially elevated in the tissue from the Zif268 knock-out mice (Fig. 7c,d).
Discussion
We report here the novel concept that Zif268, which is induced during many different forms of neuronal plasticity, regulates the expression of a subset of proteasome genes.
The rigorous statistical analysis used for our microarray analysis, with tight control of the false positive rate, ensures that the potential Zif268 targets identified can be viewed with considerable confidence. We confirmed altered expression of four of the genes identified in Zif268-transfected cells by RT-PCR. The magnitude of the change detected by RT-PCR was very similar to that detected by the microarray analysis. Consistent with the microarray analysis, these genes were all downregulated by Zif268. When we assessed the level of expression of the psmb9, psme2, Tap1, and SGK genes in CNS tissue from Zif268 knock-out mice, we found that the mRNA levels were all increased relative to wild-type mice. This is very strong evidence confirming the conclusions from the in vitro cells: that Zif268 is involved in suppressing the transcription of these genes. The results are striking, because these mice exhibit learning and memory deficits, yet this is the first demonstration of genes that show altered CNS expression as a result of the functional deletion of the Zif268 gene.
The general concept of transcriptional suppression by Zif268 is not unprecedented. In fact, the recent data from peripheral cells suggest that a major role of Zif268 is in suppression of gene expression (Cao et al., 1993; Beckmann and Wilce, 1997; Dinkel et al., 1997; Thottassery et al., 1999; Fukada and Tonks, 2001; Davis et al., 2003; Wang et al., 2005). The proteasome-related genes monitored in this study therefore join a growing list of genes with expression that can be suppressed by Zif268.
Analysis of the upstream genomic sequence of the genes identified revealed many consensus Zif268 binding sites within the proximal promoter regions, clustering immediately upstream of the TSS, as has been noted for EREs in well characterized Zif268 target genes. Proteins of the NFκB/Rel family and Sp1 have been implicated in the regulation of proteasome genes in general and the SGK, Tap1, and psmb9 genes in particular. A significant proportion of the proteasome genes showed the presence of predicted motifs containing NFκB/Rel, Sp1, and Zif268 binding sites in close proximity. These modules are extremely unlikely to occur by chance, and hence Zif268 may modulate the transcriptional response of these genes to Sp1 or NFκB/Rel, as has been noted for some other genes (Ebert et al., 1994; Cogswell et al., 1997; Srivastava et al., 1998; Chapman and Perkins, 2000; Fukada and Tonks, 2001; Guillemot et al., 2001; Davis et al., 2003; Tan et al., 2003).
The ability of Zif268 directly to modulate the activity of the psmb9, Tap1, and SGK promoters was assessed. Consistent with the other evidence reported here, the activity of all three promoter constructs was decreased by the presence of transfected Zif268.
We also tested whether these genes were modulated in a well characterized model of neuronal plasticity: dopamine antagonist-induced plasticity in the striatum. We found that all three genes were significantly modulated in striatal neurons by haloperidol administration in vivo. Interestingly, although psmb9 and Tap1 mRNAs were downregulated, SGK expression was upregulated by haloperidol treatment. However, it has become clear that, in non-neuronal cells, Zif268 target genes can be downregulated by Zif268 in some cell types and upregulated in others (Beckmann and Wilce, 1997; Copertino et al., 1997; Huang et al., 1997). A dramatic example of this is the upregulation of expression of the CBP and p300 genes in prostate cells via Zif268 after one stimulus (serum), and downregulation via Zif268 after another stimulus (UV) (Yu et al., 2004). This phenomenon is believed to reflect cell-specific or stimulus-specific interactions of Zif268 with other DNA-binding proteins. A similar phenomenon is observed with other TFs such as cAMP response element-binding protein, in which different patterns of target gene induction are observed in different situations (Zhang et al., 2005). Our results provide strong evidence that the repertoire of the downstream effects of Zif268 induction will vary in different situations according to some other factor (e.g., the presence or absence of other TFs activated in parallel with Zif268). However, increased expression of striatal SGK has also been noted in other paradigms involving Zif268 induction, the delayed response to amphetamine treatment (Gonzalez-Nicolini and McGinty, 2002) and genetic deletion of the dopamine transporter (Yao et al., 2004), suggesting that upregulation of SGK after Zif268 induction may be common in striatal neurons.
Evidence implicates both SGK and Tap1 in synaptic plasticity. SGK protects synaptic glutamate transporters from proteasomal degradation (Boehmer et al., 2003, 2004; Schniepp et al., 2004), and altered glutamate transporter levels have been suggested to contribute to LTP and memory (Levenson et al., 2002; Shen and Linden, 2005). SGK can reportedly be either upregulated or downregulated in the rodent hippocampus after performance of learning tasks (Donahue et al., 2002; Tsai et al., 2002), and it is proposed that enhanced SGK expression facilitates memory consolidation (Tsai et al., 2002). The oligopeptides generated by proteasomal protein digestion are transported into the endoplasmic reticulum via the antigen transporter complex (Tap1, Tap2, and tapasin), which is physically associated with the proteasome in many cell types (Hwang et al., 2001). Intriguingly, targeted deletion of the Tap1 gene in mice contributes to deficits in synaptic plasticity (Huh et al., 2000). Hence, in both cases, there is support for the hypothesis that regulation of these genes via Zif268 may contribute to synaptic plasticity. The consequences of reduced psmb9 expression for neuronal function are not clear, and it remains to be determined whether altered expression of psmb9 directly affects plasticity.
Our results showing regulation of proteasome gene expression by Zif268 are consistent with the evolving concept that proteasome activity is important for neuronal plasticity. The precise role of the proteasome is controversial. On the one hand, proteasome inhibitors attenuate learning and memory (Hegde et al., 1997; Chain et al., 1999; Lopez-Salon et al., 2002) and suppress functional plasticity (Moss et al., 2003), whereas compromised proteasome function has been linked with aging (Keller et al., 2000) and with memory impairment in humans (Cooper et al., 2004). This suggests a facilitatory role for the proteasome in plasticity. In contrast, proteasome inhibition enhances neurite outgrowth, presynaptic neurotransitter release, and postsynaptic glutamate receptor sensitivity, consistent with the proteasome acting to suppress plasticity (Obin et al., 1999; Speese et al., 2003; Zhao et al., 2003; Juo and Kaplan, 2004). Synaptic plasticity probably depends on a precise balance between the synthesis of some proteins and the degradation of others, as has been noted for the guidance of growth cones in response to environmental cues (Campbell and Holt, 2001). Regulation of proteasome genes in the CNS by Zif268 may allow long-lasting but potentially subtle alterations in the activity of the proteasome.
Sustained alterations in proteasome gene expression have been associated with long-term CNS plasticity. Altered expression of a number of proteasome genes, in particular psmb9, psme1, and SGK, has been observed in a variety of plasticity models (El-Khodor et al., 2001; Becker et al., 2003; Blalock et al., 2003; Cirelli et al., 2004). In addition, reduction of the chymotrypsin activity of the proteasome, as would be predicted with reduced psmb9 levels, has been specifically linked with the modulation of neuronal morphology (Fenteany and Schreiber, 1996). Thus, the regulation of proteasome gene expression, and particularly the psmb9 and psme1 genes, is likely to be of major importance for CNS function. A speculative possibility would be that, while interferon and related hormones regulate the expression of inducible proteasome subunits in the immune system, this role is assumed by glutamatergic activity in the CNS, via TFs including Zif268.
Induction or repression of the proteasome subunits psmb8, psmb9, psme1, and psme2 alters the activity of the proteasome (Groettrup et al., 1995; Harris et al., 2001). Increased Zif268 expression suppressed overall proteasome function in PC12 cells. The degree of inhibition was slight compared with the reductions in psmb9 and psme2 mRNA. However, we observed high basal proteasome activity in PC12 cells, probably because of the relatively high expression of other catalytic β subunits such as psmb3, psmb5, and psmb6 (Fig. 1). Hence, the substantial Zif268-mediated reduction in psmb9 and psme2 expression might not greatly affect overall proteasome activity (Oberdorf et al., 2001; Kazi et al., 2003; Berkers et al., 2005). Nevertheless, the results indicate that Zif268 induction suppresses proteasome activity, with the degree of suppression probably dependent on the level of expression of other catalytic proteasome subunits in the cell at that time.
Conversely, the absence of Zif268 in the gene-targeted mice was associated with substantially elevated cerebral cortex proteasome activity (Fig. 7). In the immune system, transcriptional induction of the psmb8, psmb9, psme1, and psme2 genes enhances proteasome activity, in the absence of any change in expression of the constitutive β catalytic or 19S subunits (Fruh et al., 1994; Yang et al., 1995; Hisamatsu et al., 1996). It is believed that the psmb8 and psmb9 subunits displace the corresponding constitutive β subunits, whereas the psme1 and psme2 subunits compete reversibly with the existing constitutive 19S complex. Thus, elevated proteasome activity after increased expression of the psmb8, psmb9, psme1, and psme2 subset of proteasome subunit genes would be predicted based on extrapolations from the immunoproteasome to the neuronal proteasome. Our data from the Zif268 knock-out mice confirm the importance of Zif268 for the regulation of CNS proteasome activity. They also raise the interesting possibility that abnormal proteasome activity may contribute to the impairment of late-phase LTP and the deficits in behavioral tests of learning and memory that are observed in these mice (Jones et al., 2001).
Overall, our discovery that Zif268 regulates the transcription of proteasome-related genes and hence modulates the activity of the proteasome reveals a new and previously unsuspected dimension to the mechanisms of neuronal plasticity. The regulation of proteasome function at multiple levels provides a new insight into the mechanisms sustaining the late phase of neuronal plasticity and focusses attention particularly on protein degradation and subsequent modification of synaptic transmission as underlying these enduring changes.
Footnotes
This work was supported by the Biotechnology and Biological Sciences Research Council. We thank Dr. Paul Burr (BioBest Laboratories, Edinburgh, UK) for advice in the early stages of this project; Dr. G. Eric Blair (University of Leeds, Leeds, UK) and Prof. Gerald Thiel (University of the Saarland Medical Center, Homburg, Germany) for the gift of the Tap1/psmb9 promoter construct and the pZif268 expression vector, respectively; the staff at the Sir Henry Wellcome Functional Genomics Facility (University of Glasgow, Glasgow, UK) for assistance with the microarray processing and analysis; and Prof. P. Charnay, Dr. P. Topilko, and Dr. M. Desmarquet-Trin-Dinh (Institut National de la Santé et de la Recherche Médicale U368, Paris, France) for the gift of the Zif268 knock-out mice. Jie Zang performed some of the proteasome assays.
References
- Bahouth SW, Beauchamp MJ, Vu KN (2002). Reciprocal regulation of beta 1-adrenergic receptor gene transcription by Sp1 and early growth response gene 1: induction of EGR-1 inhibits the expression of the beta 1-adrenergic receptor gene. Mol Pharmacol 61:379–390. [DOI] [PubMed] [Google Scholar]
- Becker AJ, Chen J, Zien A, Sochivko D, Normann S, Schramm J, Elger CE, Wiestler OD, Blumcke I (2003). Correlated stage- and subfield-associated hippocampal gene expression patterns in experimental and human temporal lobe epilepsy. Eur J Neurosci 18:2792–2802. [DOI] [PubMed] [Google Scholar]
- Beckmann AM, Wilce PA (1997). EGR transcription factors in the nervous system. Neurochem Int 31:477–510. [DOI] [PubMed] [Google Scholar]
- Berkers CR, Verdoes M, Lichtman E, Fiebiger E, Kessler BM, Anderson KC, Ploegh HL, Ovaa H, Galardy PJ (2005). Activity probe for in vivo profiling of the specificity of proteasome inhibitor bortezomib. Nat Methods 2:357–362. [DOI] [PubMed] [Google Scholar]
- Blalock EM, Chen K-C, Sharrow K, Herman JP, Porter NM, Foster TC, Landfield PW (2003). Gene microarrays in hippocampal aging: statistical profiling identifies novel processes correlated with cognitive impairment. J Neurosci 23:3807–3819. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boehmer C, Henke G, Schniepp R, Palmada M, Rothstein JD, Broer S, Lang F (2003). Regulation of the glutamate transporter EAAT1 by the ubiquitin ligase Nedd4–2 and SGK1/3 and protein kinase B. J Neurochem 86:1181–1188. [DOI] [PubMed] [Google Scholar]
- Boehmer C, Philippin M, Rajamanickam J, Mack A, Broer S, Palmada M, Lang F (2004). Stimulation of the EAAT4 glutamate transporter by SGK protein kinase isoforms and PKB. Biochem Biophys Res Commun 324:1242–1248. [DOI] [PubMed] [Google Scholar]
- Calabresi P, Saiardi A, Pisani A, Baik J-H, Centonze D, Mercuri NB, Bernardi G, Borrelli E (1997). Abnormal synaptic plasticity in the striatum of mice lacking dopamine D2 receptors. J Neurosci 17:4536–4544. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Campbell DS, Holt CE (2001). Chemotropic responses of retinal growth cones mediated by rapid local protein synthesis and degradation. Neuron 32:1013–1026. [DOI] [PubMed] [Google Scholar]
- Cao X, Mahendran R, Guy G, Tan Y (1993). Detection and characterization of cellular EGR-1 binding to its recognition site. J Biol Chem 268:16949–16957. [PubMed] [Google Scholar]
- Cascio P, Call M, Petre BM, Walz T, Goldberg AL (2002). Properties of the hybrid form of the 26S proteasome containing both 19S and PA28 complexes. EMBO J 21:2636–2645. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cepeda C, Hurst RS, Altemus KL, Flores-Hernandez J, Calvert CR, Jokel ES, Grandy DK, Low MJ, Rubinstein M, Ariano MA, Levine MS (2001). Facilitated glutamatergic transmission in the striatum of D2 dopamine receptor-deficient mice. J Neurophysiol 85:659–670. [DOI] [PubMed] [Google Scholar]
- Chain DG, Schwartz JH, Hegde AN (1999). Ubiquitin-mediated proteolysis in learning and memory. Mol Neurobiol 20:125–142. [DOI] [PubMed] [Google Scholar]
- Chapman NR, Perkins ND (2000). Inhibition of the RelA (p65) NFkB subunit by egr-1. J Biol Chem 275:4719–4725. [DOI] [PubMed] [Google Scholar]
- Chatterjee-Kishore M, Kishore R, Hicklin DJ, Marincola FM, Ferrone S (1998). Different requirements for signal transducer and activator of transcription 1alpha and interferon regulatory factor 1 in the regulation of low molecular mass polypeptide 2 and transporter associated with antigen processing 1 gene expression. J Biol Chem 273:16177–16183. [DOI] [PubMed] [Google Scholar]
- Chowdhary R, Ali RA, Albig W, Doenecke D, Bajic VB (2005). Promoter modeling: the case study of mammalian histone promoters. Bioinformatics 21:2623–2628. [DOI] [PubMed] [Google Scholar]
- Cirelli C, Gutierrez CM, Tononi G (2004). Extensive and divergent effects of sleep and wakefulness on brain gene expression. Neuron 41:35–43. [DOI] [PubMed] [Google Scholar]
- Cogswell PC, Mayo MW, Baldwin AS Jr (1997). Involvement of Egr-1/RelA synergy in distinguishing T cell activation from tumor necrosis factor-alpha-induced NF-kappa B1 transcription. J Exp Med 185:491–498. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cole AJ, Saffen DW, Baraban JM, Worley PF (1989). Rapid increase of an immediate-early gene mRNA in hippocampal neurons by NMDA receptor activation. Nature 340:474–476. [DOI] [PubMed] [Google Scholar]
- Colledge M, Snyder EM, Crozier RA, Soderling JA, Jin Y, Langeberg LK, Lu H, Bear MF, Scott JD (2003). Ubiquitination regulates PSD-95 degradation and AMPA receptor surface expression. Neuron 40:595–607. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Conway A-M, James AB, O’Kane EM, Rakhit S, Morris BJ (2004). Regulation of myosin light chain phosphorylation by RhoB in neuronal cells. Exp Cell Res 300:35–42. [DOI] [PubMed] [Google Scholar]
- Cooper EM, Hudson AW, Amos J, Wagstaff J, Howley PM (2004). Biochemical analysis of Angelman syndrome-associated mutations in the E3 ubiquitin ligase E6-associated protein. J Biol Chem 279:41208–41217. [DOI] [PubMed] [Google Scholar]
- Copertino DW, Edelman GM, Jones FS (1997). Multiple promoter elements differentially regulate the expression of the mouse tenascin gene. Proc Natl Acad Sci USA 94:1846–1851. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Davis W Jr, Chen ZJ, Ile KE, Tew KD (2003). Reciprocal regulation of expression of the human adenosine 5′-triphosphate binding cassette, sub-family A, transporter 2 (ABCA2) promoter by the early growth response-1 (EGR-1) and Sp-family transcription factors. Nucleic Acids Res 31:1097–1107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Diaz-Hernandez M, Hernandez F, Martin-Aparicio E, Gomez-Ramos P, Moran MA, Casano JG, Ferrer I, Avila J, Lucas JJ (2003). Neuronal induction of the immunoproteasome in Huntington’s disease. J Neurosci 23:11653–11661. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ding Q, Keller JN (2001). Proteasomes and proteasome inhibition in the central nervous system. Free Radic Biol Med 31:574–584. [DOI] [PubMed] [Google Scholar]
- Dinkel A, Aicher W, Haas C, Zipfel P, Peter H, Eibel H (1997). Transcription factor Egr-1 activity down-regulates Fas and CD23 expression in B cells. J Immunol 159:2678–2684. [PubMed] [Google Scholar]
- Donahue CP, Jensen RV, Ochiishi T, Eisenstein I, Zhao M, Shors T, Kosik KS (2002). Transcriptional profiling reveals regulated genes in the hippocampus during memory formation. Hippocampus 12:821–833. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ebert SN, Balt SL, Hunter JPB, Gashler A, Sukhatme V, Wong DL (1994). Egr-1 activation of rat adrenal phenylethanolamine N-methyltransferase gene. J Biol Chem 269:20885–20898. [PubMed] [Google Scholar]
- Ehlers MD (2003). Activity level controls postsynaptic composition and signaling via the ubiquitin-proteasome system. Nat Neurosci 6:231–242. [DOI] [PubMed] [Google Scholar]
- El-Khodor BF, Kholodilov NG, Yarygina O, Burke RE (2001). The expression of mRNAs for the proteasome complex is developmentally regulated in the rat mesencephalon. Dev Brain Res 129:47–56. [DOI] [PubMed] [Google Scholar]
- Fenteany G, Schreiber SL (1996). Specific inhibition of the chymotrypsin-like activity of the proteasome induces a bipolar morphology in neuroblastoma cells. Chem Biol 3:905–912. [DOI] [PubMed] [Google Scholar]
- Fruh K, Gossen M, Wang K, Bujard H, Peterson P, Yang Y (1994). Displacement of housekeeping proteasome subunits by MHC-encoded LMPs: a newly discovered mechanism for modulating the multicatalytic proteinase complex. EMBO J 13:3236–3244. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fukada T, Tonks NK (2001). The reciprocal role of Egr-1 and Sp family proteins in regulation of the PTP1B promoter in response to the p210 Bcr-Abl oncoprotein-tyrosine kinase. J Biol Chem 276:25512–25519. [DOI] [PubMed] [Google Scholar]
- Gonzalez-Nicolini V, McGinty JF (2002). Gene expression profile from the striatum of amphetamine-treated rats: a cDNA array and in situ hybridization histochemical study. Brain Res Gene Expr Patterns 1:193–198. [DOI] [PubMed] [Google Scholar]
- Groettrup M, Ruppert T, Kuehn L, Seeger M, Standera S, Koszinowski U, Kloetzel PM (1995). The interferon-gamma-inducible 11 S regulator (PA28) and the LMP2/LMP7 subunits govern the peptide production by the 20 S proteasome in vitro. J Biol Chem 270:23808–23815. [DOI] [PubMed] [Google Scholar]
- Groot M, Boxer LM, Thiel G (2000). Nerve growth factor- and epidermal growth factor-regulated gene transcription in PC12 pheochromocytoma and INS-1 insulinoma cells. Eur J Cell Biol 79:924–935. [DOI] [PubMed] [Google Scholar]
- Guillemot L, Levy A, Raymondjean M, Rothhut B (2001). Angiotensin II-induced transcriptional activation of the cyclin D1 gene is mediated by Egr-1 in CHO-AT1A cells. J Biol Chem 276:39394–39403. [DOI] [PubMed] [Google Scholar]
- Harris JL, Alper PB, Li J, Rechsteiner M, Backes BJ (2001). Substrate specificity of the human proteasome. Chem Biol 8:1131–1141. [DOI] [PubMed] [Google Scholar]
- Hegde AN, Inokuchi K, Pei W, Casadio A, Ghirardi M, Chain DG, Martin KC, Kandel ER, Schwartz JH (1997). Ubiquitin C-terminal hydrolase is an immediate-early gene essential for long-term facilitation in Aplysia. Cell 89:115–126. [DOI] [PubMed] [Google Scholar]
- Hisamatsu H, Shimbara N, Saito Y, Kristensen P, Hendil K, Fujiwara T, Takahashi E, Tanahashi N, Tamura T, Ichihara A, Tanaka K (1996). Newly identified pair of proteasomal subunits regulated reciprocally by interferon gamma. J Exp Med 183:1807–1816. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang R-P, Fan Y, Ni Z, Mercola D, Adamson ED (1997). Reciprocal modulation between Sp1 and Egr1. J Cell Biochem 66:489–499. [PubMed] [Google Scholar]
- Huh GS, Boulanger LM, Du H, Riquelme PA, Brotz TM, Shatz CJ (2000). Functional requirement for class I MHC in CNS development and plasticity. Science 290:2155–2159. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hwang L-Y, Lieu PT, Peterson PA, Yang Y (2001). Functional regulation of immunoproteasomes and transporter associated with antigen processing. Immunol Res 24:245–272. [DOI] [PubMed] [Google Scholar]
- James AB, Conway A-M, Thiel G, Morris BJ (2004). Egr1-mediated transcriptional modulation of neuronal target genes: permissive effect of forskolin via cAMP. Cell Signal 16:1355–1362. [DOI] [PubMed] [Google Scholar]
- James AB, Conway A-M, Morris BJ (2005). Genomic profiling of the neuronal target genes of the plasticity-related transcription factor—Zif268. J Neurochem 95:796–810. [DOI] [PubMed] [Google Scholar]
- Jarvis ED, Scharff C, Grossman MR, Ramos JA, Nottebohm F (1998). For whom the bird sings: context-dependent gene expression. Neuron 21:775–788. [DOI] [PubMed] [Google Scholar]
- Jones MW, Errington ML, French PJ, Fine A, Bliss TVP, Garel S, Charnay P, Bozon B, Laroche S, Davis S (2001). A requirement for the immediate early gene Zif268 in the expression of late LTP and long-term memories. Nat Neurosci 4:289–296. [DOI] [PubMed] [Google Scholar]
- Juo P, Kaplan JM (2004). The anaphase-promoting complex regulates the abundance of GLR-1 glutamate receptors in the ventral nerve cord of C. elegans Curr Biol 14:2057–2062. [DOI] [PubMed] [Google Scholar]
- Kaczmarek L, Zangenehpour S, Chaudhuri A (1999). Sensory regulation of immediate-early genes c-fos and zif268 in monkey visual cortex at birth and throughout the critical period. Cereb Cortex 9:179–187. [DOI] [PubMed] [Google Scholar]
- Kamalakaran S, Radhakrishnan SK, Beck WT (2005). Identification of estrogen-responsive genes using a genome-wide analysis of promoter elements for transcription factor binding sites. J Biol Chem 280:21491–21497. [DOI] [PubMed] [Google Scholar]
- Kazi A, Daniel KG, Smith DM, Kumar NB, Dou QP (2003). Inhibition of the proteasome activity, a novel mechanism associated with the tumor cell apoptosis-inducing ability of genistein. Biochem Pharmacol 66:965–976. [DOI] [PubMed] [Google Scholar]
- Keefe KA, Gerfen CR (1995). D1–D2 dopamine receptor synergy in striatum: effects of intrastriatal infusions of dopamine agonists and antagonists on immediate early gene expression. Neuroscience 66:903–913. [DOI] [PubMed] [Google Scholar]
- Keller JN, Hanni KB, Markesbery WR (2000). Possible involvement of proteasome inhibition in aging: implications for oxidative stress. Mech Ageing Dev 113:61–70. [DOI] [PubMed] [Google Scholar]
- Kel-Margoulis OV, Tchekmenev D, Kel AE, Goessling E, Hornischer K, Lewicki-Potapov B, Wingender E (2003). Composition-sensitive analysis of the human genome for regulatory signals. In Silico Biol 3:145–171. [PubMed] [Google Scholar]
- Levenson J, Weeber E, Selcher JC, Kategaya LS, Sweatt JD, Eskin A (2002). Long-term potentiation and contextual fear conditioning increase neuronal glutamate uptake. Nat Neurosci 5:155–161. [DOI] [PubMed] [Google Scholar]
- Liu C, Calogero A, Ragona G, Adamson E, Mercola D (1996). EGR-1, the reluctant suppression factor. Crit Rev Oncog 7:101–125. [PubMed] [Google Scholar]
- Lopez-Salon M, Alonso M, Vianna MRM, Viola H, Mello e Souza T, Izquierdo I, Pasquini JM, Medina JH (2002). The ubiquitin-proteasome cascade is required for mammalian long-term memory formation. Eur J Neurosci 14:1820–1826. [DOI] [PubMed] [Google Scholar]
- Marques L, Brucet M, Lloberas J, Celada A (2004). STAT1 regulates lipopolysaccharide- and TNF-a-dependent expression of transporter associated with antigen processing 1 and low molecular mass polypeptide 2 genes in macrophages by distinct mechanisms. J Immunol 173:1103–1110. [DOI] [PubMed] [Google Scholar]
- Mello CV, Velho TAF, Pinaud R (2004). Song-induced gene expression: a window on song auditory processing and perception. Ann NY Acad Sci 1016:263–281. [DOI] [PubMed] [Google Scholar]
- Mengual E, Arizti P, Rodrigo J, Gimenez-Amaya JM, Castano JG (1996). Immunohistochemical distribution and electron microscopic subcellular localization of the proteasome in the rat CNS. J Neurosci 16:6331–6341. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Morris BJ (1995). Stimulation of immediate-early gene expression in striatal neurones by nitric oxide. J Biol Chem 270:24740–24744. [PubMed] [Google Scholar]
- Morris BJ (2004). Neuronal plasticity. In: Molecular biology of the neuron (Davies RW, Morris BJ, eds) , Ed 2 pp. 357–386. Oxford: Oxford UP.
- Moss A, Blackburn-Munro G, Garry EM, Blakemore JA, Dickinson T, Resie R, Mitchell R, Fleetwood-Walker SM (2003). A role of the ubiquitin–proteasome system in neuropathic pain. J Neurosci 22:1363–1372. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nguyen TV, Kosofsky BE, Birnbaum R, Cohen BM, Hyman SE (1992). Differential expression of c-fos and zif268 in rat striatum after haloperidol, clozapine, and amphetamine. Proc Natl Acad Sci USA 89:4270–4274. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Noda C, Tanahashi N, Shimbara N, Hendil KB, Tanaka K (2000). Tissue distribution of constitutive proteasomes, immunoproteasomes, and PA28 in rats. Biochem Biophys Res Commun 277:348–354. [DOI] [PubMed] [Google Scholar]
- Oberdorf J, Carlson EJ, Skach WR (2001). Redundancy of mammalian proteasome beta subunit function during endoplasmic reticulum associated degradation. Biochemistry 40:13397–13405. [DOI] [PubMed] [Google Scholar]
- Obin M, Mesco E, Gong X, Haas AL, Joseph J, Taylor A (1999). Neurite outgrowth in PC12 cells. Distinguishing the roles of ubiquitylation and ubiquitin-dependent proteolysis. J Biol Chem 274:11789–11795. [DOI] [PubMed] [Google Scholar]
- Pak DTS, Sheng M (2003). Targeted protein degradation and synapse remodelling by an inducible protein kinase. Science 302:1368–1373. [DOI] [PubMed] [Google Scholar]
- Proffitt JA, Blair GE (1997). The MHC-encoded TAP1/LMP2 bidirectional promoter is down-regulated in highly oncogenic adenovirus type 12 transformed cells. FEBS Lett 400:141–144. [DOI] [PubMed] [Google Scholar]
- Quandt K, Frech K, Karas H, Wingender E, Werner T (1995). Matind and Matinspector—new fast and versatile tools for detection of consensus matches in nucleotide-sequence data. Nucleic Acids Res 23:4878–4884. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rezvani K, Mee S, Dawson S, McIlhinney J, Fujita J, Mayer RJ (2003). Proteasomal interactors control activities as diverse as the cell cycle and glutaminergic neurotransmission. Biochem Soc Trans 31:470–473. [DOI] [PubMed] [Google Scholar]
- Richardson CL, Tate WP, Mason SP, Lawlow PA, Dragunow M, Abraham WC (1992). Correlation betwen the induction of an immediate-early gene, zif/268, and long-term potentiation. Brain Res 580:147–154. [DOI] [PubMed] [Google Scholar]
- Schniepp R, Kohler K, Ladewig T, Guenther E, Henke G, Palmada M, Boehmer C, Rothstein JD, Broer S, Lang F (2004). Retinal colocalization and in vitro interaction of the glutamate receptor EAAT3 and the serum- and glucocorticoid-inducible kinase SGK1. Invest Ophthalmol Vis Sci 45:1442–1449. [DOI] [PubMed] [Google Scholar]
- Shen Y, Linden DJ (2005). Long-term potentiation of neuronal glutamate transporters. Neuron 46:715–722. [DOI] [PubMed] [Google Scholar]
- Simpson CS, Morris BJ (1994). Haloperidol and fluphenazine induce junB gene expression in rat striatum and nucleus accumbens. J Neurochem 63:1955–1961. [DOI] [PubMed] [Google Scholar]
- Simpson CS, Morris BJ (2000). Regulation of neuronal cell adhesion molecule (NCAM) expression by NF-kB. J Biol Chem 275:16879–16884. [DOI] [PubMed] [Google Scholar]
- Speese SD, Trotta N, Rodesch CK, Aravamudan B, Broadie K (2003). The ubiquitin proteasome system acutely regulates presynaptic protein turnover and synaptic efficacy. Curr Biol 13:899–910. [DOI] [PubMed] [Google Scholar]
- Srivastava S, Weitzmann MN, Kimble RB, Rizzo M, Zahner M, Milbrandt J, Ross FP, Pacifici R (1998). Estrogen blocks M-CSF gene expression and osteoclast formation by regulating phosphorylation of egr1 and its interaction with Sp1. J Clin Invest 102:1850–1859. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tan L, Peng H, Osaki M, Choy BK, Auron PE, Sandell LJ, Goldring MB (2003). Egr-1 mediates transcriptional repression of COL2A1 promoter activity by interleukin-1beta. J Biol Chem 278:17688–17700. [DOI] [PubMed] [Google Scholar]
- Thottassery JV, Sun D, Zambetti GP, Troutman A, Sukhatme VP, Schuetz EG, Schuetz JD (1999). Sp1 and Egr-1 have opposing effects on the regulation of the rat Pgp2/mdr1b gene. J Biol Chem 274:3199–3206. [DOI] [PubMed] [Google Scholar]
- Topilko P, Schneider-Maunoury S, Levi G, Trembleau A, Gourdji D, Driancourt M-A, Rao CV, Charnay P (1998). Multiple pituitary and ovarian defects in Krox-24 (NGFI-A, Egr-1)-targeted mice. Mol Endocrinol 12:107–122. [DOI] [PubMed] [Google Scholar]
- Tsai KJ, Chen SK, Ma YL, Hsu WL, Lee EHY (2002). sgk, a primary glucocorticoid-induced gene, facilitates memory consolidation of spatial learning in rats. Proc Natl Acad Sci USA 99:3990–3995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tullai JW, Schaffer ME, Mullenbrock S, Kasif S, Cooper GM (2004). Identification of transcription factor binding sites upstream of human genes regulated by the phosphatidylinositol 3-kinase and MEK/ERK signaling pathways. J Biol Chem 279:20167–20177. [DOI] [PubMed] [Google Scholar]
- Tusher VG, Tibshirani R, Chu G (2001). Significance analysis of microarrays applied to the ionizing radiation response. Proc Natl Acad Sci USA 98:5116–5121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Virolle T, Krones-Herzig A, Baron V, De Gregorio G, Adamson ED, Mercola D (2003). Egr1 promotes growth and survival of prostate cancer cells. Identification of novel Egr1 target genes. J Biol Chem 278:11802–11810. [DOI] [PubMed] [Google Scholar]
- Waldegger S, Erdel M, Nagl UO, Barth P, Raber G, Steuer S, Utermann G, Paulmichl M, Lang F (1998). Genomic organization and chromosomal localization of the human SGK protein kinase gene. Genomics 51:299–302. [DOI] [PubMed] [Google Scholar]
- Wang C, Dostanic S, Servant N, Chalifour LE (2005). Egr-1 negatively regulates expression of the sodium-calcium exchanger-1 in cardiomyocytes in vitro and in vivo. Cardiovasc Res 65:187–194. [DOI] [PubMed] [Google Scholar]
- Webster M, Goya L, Ge Y, Maiyar A, Firestone G (1993). Characterization of sgk, a novel member of the serine/threonine protein kinase gene family which is transcriptionally induced by glucocorticoids and serum. Mol Cell Biol 13:2031–2040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Whitehouse AS, Tisdale MJ (2003). Increased expression of the ubiquitin-proteasome pathway in murine myotubes by proteolysis-inducing factor (PIF) is associated with activation of the transcription factor NF-kappaB. Br J Cancer 89:1116–1122. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wisden W, Errington ML, Williams S, Dunnett SB, Waters C, Hitchcock D, Evan G, Bliss TVP, Hunt SP (1990). Differential expression of immediate-early genes in the hippocampus and spinal cord. Neuron 4:603–614. [DOI] [PubMed] [Google Scholar]
- Worley PF, Bhat RV, Baraban JM, Erickson CA, Mcnaughton BL, Barnes CA (1993). Thresholds for synaptic activation of transcription factors in hippocampus: correlation with long-term enhancement. J Neurosci 13:4776–4786. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wright KL, White LC, Kelly A, Beck S, Trowsdale J, Ting J-Y (1995). Coordinate regulation of the human TAP1 and LMP2 genes from a shared bidirectional promoter. J Exp Med 181:1459–1471. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xie X, Lu J, Kulbokas EJ, Golub TR, Mootha V, Lindblad-Toh K, Lander ES, Kellis M (2005). Systematic discovery of regulatory motifs in human promoters and 3′ UTRs by comparison of several mammals. Nature 434:338–345. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang Y, Früh K, Ahn K, Peterson PA (1995). In vivo assembly of the proteasomal complexes, implications for antigen processing. J Biol Chem 270:27687–27694. [DOI] [PubMed] [Google Scholar]
- Yao WD, Gainetdinov RR, Arbuckle MI, Sotnikova TD, Cyr M, Beaulieu JM, Torres GE, Grant SG, Caron MG (2004). Identification of PSD-95 as a regulator of dopamine-mediated synaptic and behavioral plasticity. Neuron 41:625–638. [DOI] [PubMed] [Google Scholar]
- Yu J, de Belle I, Liang H, Adamson ED (2004). Coactivating factors p300 and CBP are transcriptionally crossregulated by Egr1 in prostate cells, leading to divergent responses. Mol Cell 15:83–94. [DOI] [PubMed] [Google Scholar]
- Zhang X, Liu Y (2003). Suppression of HGF receptor gene expression by oxidative stress is mediated through the interplay between Sp1 and Egr-1. Am J Physiol Renal Physiol 284:F1216–F1225. [DOI] [PubMed] [Google Scholar]
- Zhang X, Odom DT, Koo S-H, Conkright MD, Canettieri G, Best J, Chen H, Jenner R, Herbolsheimer E, Jacobsen E, Kadam S, Ecker JR, Emerson B, Hogenesch JB, Unterman T, Young RA, Montminy M (2005). Genome-wide analysis of cAMP-response element binding protein occupancy, phosphorylation, and target gene activation in human tissues. Proc Natl Acad Sci USA 102:4459–4464. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhao Y, Hegde AN, Martin KC (2003). The ubiquitin proteasome system functions as an inhibitory constraint on synaptic strengthening. Curr Biol 13:887–898. [DOI] [PubMed] [Google Scholar]
- Zheng J, Wu J, Sun Z (2003). An approach to identify over-represented cis-elements in related sequences. Nucleic Acids Res 31:1995–2005. [DOI] [PMC free article] [PubMed] [Google Scholar]