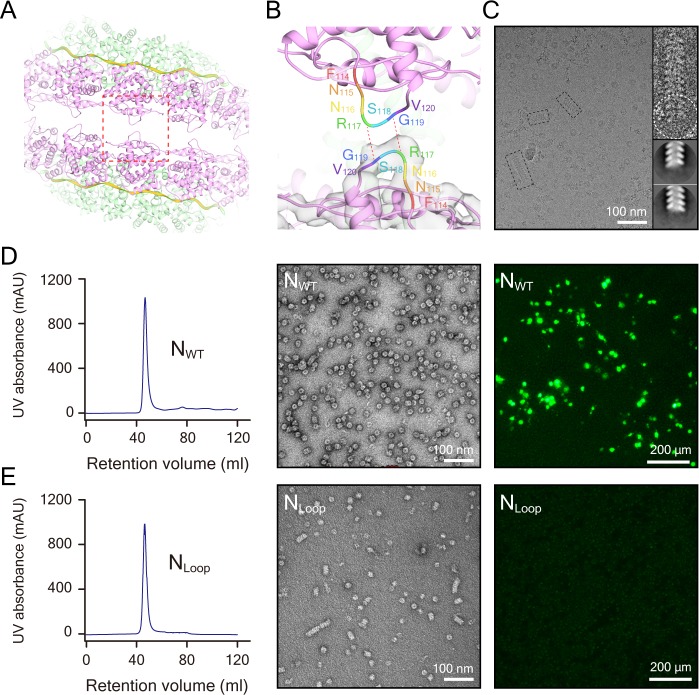
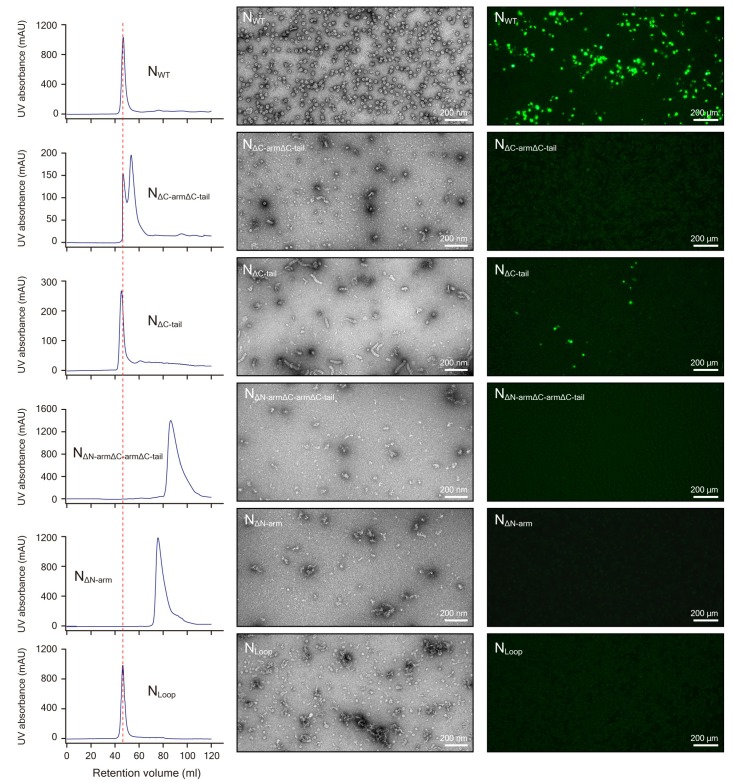
Figure 3. The clam-shaped nucleocapsid is important for the function of the viral genome.
(A) Loop pairs from the vertically adjacent N form the self-capping interface in the clam-shaped structure. Five loop pairs furthest from the seam are shown. Colors are as in Figure 1. (B) View of one loop pair of the clam-shaped structure. Seven residues (114–120) in the upper loop are labeled and the lower loop is docked into the EM density. (C) Raw micrograph of a single-headed helix from the NLoop and the 2D classification of the filament tip (circled). Zoomed-in view of selected raw filaments (examples in dashed boxes) with two typical 2D classes on the tip shown. (D) NWT was able to form double-headed filaments and functioned well in the minigenome assay. The retention volume of NWT in gel filtration chromatography was ~47 ml (left) and the negative-stain image of this fraction consisted of a clam-shaped structure and filaments was zoomed in (middle). NWT exhibited strong fluorescence signals in a minigenome assay in BSR-T7/5 cells (right). (E) The NLoop formed filaments but was not functional in a minigenome assay. The retention volume of the NLoop was ~47 ml, close to NWT (left). Negative-stain EM showed more filaments than NWT (middle). However, there was no fluorescence signal in the minigenome assay (right).