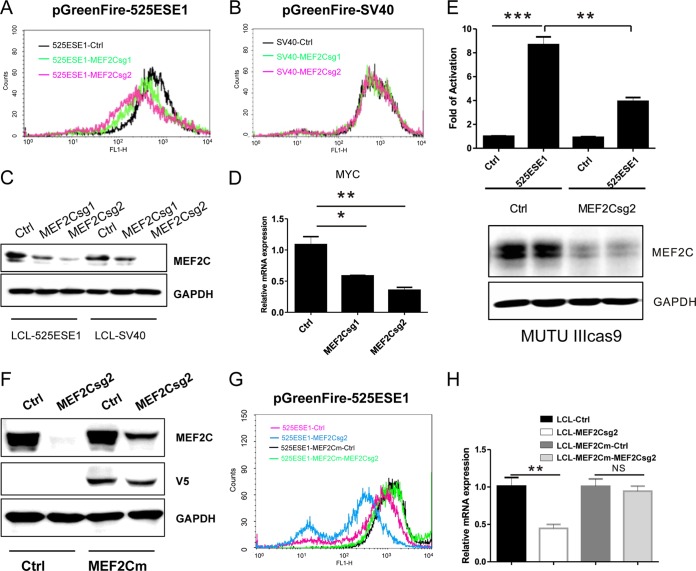
FIG 6.
MEF2C is essential for 5252ESE1 function. (A) LCLs stably expressing CAS9 and 525ESE1-driven GFP-luciferase reporter were transduced with lentiviruses expressing control sgRNA or sgRNA targeting MEF2C. After puromycin selection, GFP levels were determined by FACS. (B) LCLs stably expressing CAS9 and SV40 enhancer-driven GFP-luciferase reporter were transduced with lentiviruses expressing control sgRNA or sgRNA targeting MEF2C. GFP levels were determined by FACS. (C) MEF2C expression in LCLs expressing 525ESE1- or SV40 enhancer-driven reporters. (D) MYC expression levels determined by qRT-PCR following MEF2C knockout. The level of control sgRNA was set to 1. **, P < 0.01; *, P < 0.05. (E) MEF2C expression was first knocked out by CRISPR in MUTU III cells. 525ESE1-driven reporter or control reporter was then electroporated into these cells. Luciferase activities were normalized by Renilla luciferase. Control reporter levels were set to 1. ***, P < 0.001; **, P < 0.01. (F) V5-tagged MEF2C cDNA with the 525ESE1 sgRNA2 PAM site mutated was expressed in LCLs. MEF2C sgRNA2 efficiently knocked out endogenous MEF2C but not MEF2C from rescue cDNA. (G) 525ESE1-driven GFP levels in cells with MEF2C knockout or cells expressing rescue cDNA. MEF2C knockout efficiently repressed MYC expression. (H) In cells expressing CRISPR-resistant MEF2C, knockout did not decrease MYC expression. MYC expression levels in control sgRNA treated cells were set to 1. **, P < 0.01; NS, not significant.