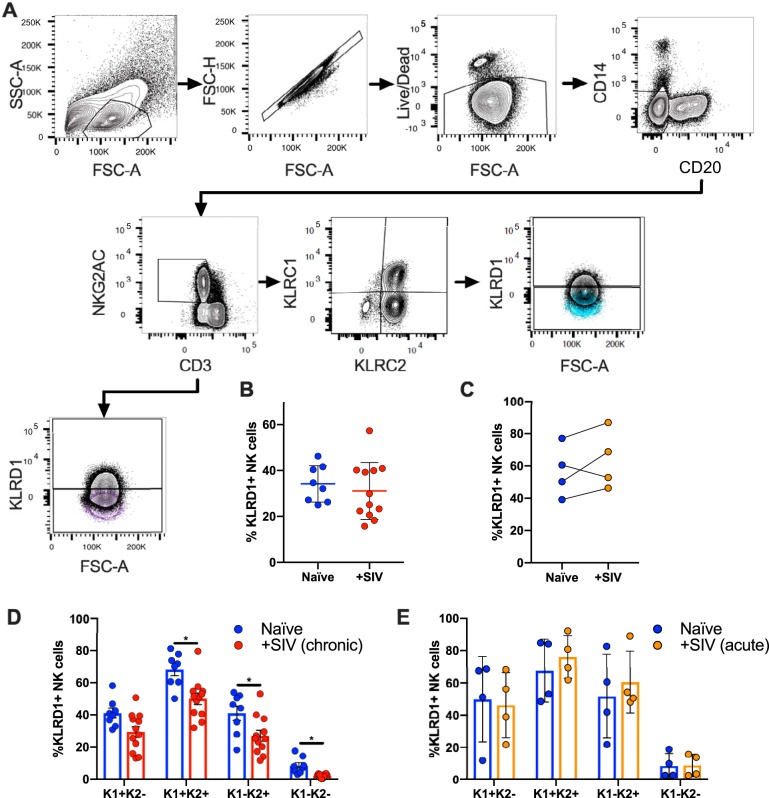
FIG 1.
Gating strategy showing identification of KLRD1+ NK cells. (A) NK cells were defined as CD20− CD14− CD3− NKG2AC+. This population was further subdivided into either KLRD1+ and KLRD1− populations (a representative FMO control is shown in purple) or first subdivided into KLRC1 and KLRC2 and then into KLRD1+ and KLRD1− populations (a representative FMO control is shown in blue). (B) Plots showing matched KLRD1+ NK cells in NKG2AC+ NK cells from naive and chronically (B) or acutely (C) SIV-infected rhesus macaques. Quantification of matched naive and chronically SIV-infected (D) or acute-infected (E) cohorts showing distribution of KLRD1+ NK cells after being subdivided into KLRC1+/− KLRC2+/− NK cell subsets. Naive samples are shown in blue, chronic SIV samples are shown in red, and acute SIV samples are shown in orange. KLRC1 was represented as K1, KLRC2 is represented as K2, and quadrant populations are represented as combinations of K1 and K2. A Wilcoxon test was used to compare different quadrant populations (*, P < 0.05).