Abstract
Flavobacterium anhuiense (previously identified as Flavobacterium johnsoniae) strain GSE09 is a volatile-producing bacterium that exhibits significant biocontrol activity against an oomycete pathogen, Phytophthora capsici, on pepper plants. Here, we report the complete genome sequence data of strain GSE09, isolated from surface-sterilized cucumber root. The genome consists of a circular 5,109,718-bp chromosome with a G + C content of 34.30%. A total of 4,138 complete coding sequences including 15 rRNA, 66 tRNA, 3 ncRNA, and 51 pseudogene sequences were retrieved. Thus, the genome sequence data of F. anhuiense GSE09 may facilitate the elucidation of many biological traits related to the biocontrol against plant pathogens.
Keywords: Biocontrol, Complete genome sequence, EggNOG analysis, Flavobacterium anhuiense, Phytophthora capsici
Specification table
| Subject area | Biology |
| More specific subject area | Microbiology and Genomics |
| Type of data | Complete genome sequence data of Flavobacterium anhuiense GSE09 |
| How data was acquired | Genome sequencing using PacBio RS II at Theragen Etex Bio Institute, Suwon, South Korea |
| Data format | Raw and analyzed data |
| Experimental factors | DNA was extracted from F. anhuiense GSE09 |
| Experimental features | Whole genome sequencing, assembly, and annotation. |
| Data source location | F. anhuiense GSE09 was isolated from the surface-sterilized root of a cucumber plant grown in a field in Gunsan, Korea. |
| Data accessibility | The genome sequence of F. anhuiense GSE09 has been deposited in DDBJ/ENA/GenBank under the accession number CP016907 (https://www.ncbi.nlm.nih.gov/nuccore/CP016907) |
| Related research | M.K. Sang, K.D. Kim, The volatile-producing Flavobacterium johnsoniae strain GSE09 shows biocontrol activity against Phytophthora capsici in pepper. J. Appl. Microbiol. 113 (2012) [1] |
Value of the data
|
1. Data
Previous studies [1], [2], [3] showed that strain GSE09, isolated from the surface-sterilized root of a cucumber (Cucumis sativus) plant grown in a field in Gunsan, South Korea in 2002, significantly inhibited infections by a soilborne oomycete pathogen, Phytophthora capsici, the causal agent of Phytophthora blight of pepper. This strain produces the volatile organic compound (VOC), 2,4-di-tert-butylphenol, which inhibits mycelial growth, sporulation, and zoospore germination of the pathogen [1,3]. In a previous study [1], strain GSE09 was identified as Flavobacterium johnsoniae; however, it has now been re-identified as Flavobacterium anhuiense based on phylogenetic analysis of the 16S rRNA gene sequences (Fig. 1).
Fig. 1.
Neighbor-joining phylogenetic tree, based on 16S rRNA gene sequences, showing the relationship between strain GSE09 (GenBank accession no. EU034657) and other species of the genus Flavobacterium. Filled circles on the branching points indicate that the corresponding nodes were also recovered in trees constructed using the maximum-likelihood and maximum-parsimony algorithms. Bootstrap values (≥70%) of 1000 analyses are shown at the branch points. Leeuwenhoekiella marinoflavo ATCC 1932T was used as an outgroup. Bar, 1 nt substitution per 100 nt of the 16S rRNA gene sequence.
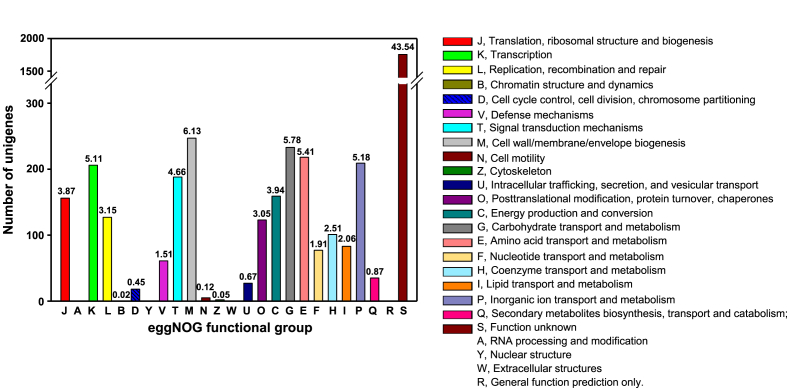
The genome of strain GSE09 consists of a circular 5,109,718-bp chromosome with a G + C content of 34.30%. In addition, 4,138 complete coding sequences (CDSs) were predicted, out of which 15 rRNA, 66 tRNA, 3 ncRNA, and 51 pseudogene sequences were retrieved. Moreover, 2,276 unigenes (56.46%) out of a total of 4,031 unigenes in GSE09 were assigned to 20 out of 24 functional groups based on the evolutionary genealogy of genes: non-supervised orthologous groups (eggNOG) analysis (Fig. 2). A total of 247 (6.13%), 233 (5.78%), and 218 (5.41%) unigenes were assigned to the ‘cell wall/membrane/envelope biogenesis’, ‘carbohydrate transport and metabolism’, and ‘amino acid transport and metabolisms’ groups, respectively; however, no unigenes were assigned to the ‘RNA processing and modification’, ‘nuclear structure’, ‘extracellular structures’, and ‘general function prediction’ groups. Furthermore, 61 unigenes were assigned to ‘defense mechanisms’ and 5 were assigned to ‘cell motility’, which may be related to the biocontrol traits of the strain (Fig. 2).
Fig. 2.
Number and percentage of Flavobacterium anhuiense strain GSE09 unigenes assigned to each functional group using the evolutionary genealogy of genes: non-supervised orthologous groups (eggNOG) analysis. The number on the bar represents the percentage of unigenes assigned to each functional group out of the total numbers of unigenes.
The genome of strain GSE09 also contained genes related to colonization (e.g., motility protein B, swarming motility protein SwrC, and gliding motility lipoprotein GldH precursor) [4], [5], [6] and antibacterial activity (e.g., thiazole synthase and polyketide cyclase/dehydrase and lipid transport) [7], [8]. Furthemore, it contained various genes related to plant defense against plant pathogens and genes related to plant growth promotion, such as ammonia, siderophores, and indole-3-acetic acid (IAA) [9], [10], [11].
2. Experimental design, materials and methods
Strain GSE09 was re-identified using phylogenetic analysis of the 16S rRNA gene sequence data (accession no. EU034657) in NCBI [1]. The analysis was performed with strain GSE09 and type strains of species belonging to the genus Flavobacterium using the molecular evolutionary genetics analysis (MEGA) version 6 software. Phylogenetic trees were constructed using neighbor-joining, maximum-likelihood, and maximum-parsimony algorithms [12], [13], [14]. The tree topology was confirmed via 1000 replications of bootstrap analysis [15].
The genomic DNA of strain GSE09, which was prepared as described previously [1], was extracted using the DNeasy Plant Mini Kit (Qiagen, Valencia, CA, USA) according to the manufacturer's protocols. Genome sequencing of the strain was performed using the PacBio RS II instrument (Pacific Bioscience, Menlo Park, CA, USA) at the Theragen Etex Bio Institute (Suwon, South Korea). Single-molecule real-time (SMRT) sequencing with PacBio RS II was conducted with SMRTbell DNA template libraries that were generated using the standard 20-kb library protocols of the PacBio SMRTbell template prep kit (Pacific Bioscience). The sequence reads generated were used for de novo assembly using the PacBio Hierarchical Genome Assembly Process version 3 (HGAP3), which were then polished using Quiver [16]. Genome annotation of strain GSE09 was performed using the Prokaryotic Genome Annotation Pipeline (PGAP) service provided by NCBI. Furthermore, the eggNOG analysis of strain GSE09 genes was conducted using CLgenomics ver. 1. 53 [17].
Acknowledgments
Jin-Ju Jeong was supported by the Global Ph.D. program through the National Research Foundation of Korea funded by the Ministry of Education (2015-034526), South Korea. Siti Sajidah was supported by the Korean Government Scholarship Program (KGSP) during her M.S. study at Korea University, Seoul, South Korea.
Conflict of interest
The authors declare that they have no known competing financial interests or personal relationships that could have appeared to influence the work reported in this paper.
References
- 1.Sang M.K., Kim K.D. The volatile-producing Flavobacterium johnsoniae strain GSE09 shows biocontrol activity against Phytophthora capsici in pepper. J. Appl. Microbiol. 2012;113:383–398. doi: 10.1111/j.1365-2672.2012.05330.x. [DOI] [PubMed] [Google Scholar]
- 2.Sang M.K., Chun S.C., Kim K.D. Biological control of Phytophthora blight of pepper by antagonistic rhizobacteria selected from a sequential screening procedure. Biol. Control. 2008;46:424–433. [Google Scholar]
- 3.Sang M.K., Kim J.D., Kim B.S., Kim K.D. Root treatment with rhizobacteria antagonistic to Phytophthora blight affects anthracnose occurrence, ripening, and yield of pepper fruit in the plastic house and field. Phytopathology. 2011;101:666–678. doi: 10.1094/PHYTO-08-10-0224. [DOI] [PubMed] [Google Scholar]
- 4.Luo Y., Cheng Y., Yi J., Zhang Z., Luo Q., Zhang D., Li Y. Complete genome sequence of industrial biocontrol strain Paenibacillus polymyxa HY96-2 and further analysis of its biocontrol mechanism. Front. Microbiol. 2018;9:1520. doi: 10.3389/fmicb.2018.01520. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Kearns D.B., Chu F., Rudner R., Losick R. Genes governing swarming in Bacillus subtilis and evidence for a phase variation mechanism controlling surface motility. Mol. Microbiol. 2004;52:357–369. doi: 10.1111/j.1365-2958.2004.03996.x. [DOI] [PubMed] [Google Scholar]
- 6.McBride M.J., Braun T.F., Brust J.L. Flavobacterium johnsoniae GldH is a lipoprotein that is required for gliding motility and chitin utilization. J. Bacteriol. 2003;185:6648–6657. doi: 10.1128/JB.185.22.6648-6657.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Kaspady M., Narayanaswamy V.K., Raju M., Rao G.K. Synthesis, antibacterial activity of 2, 4-disubstituted oxazoles and thiazoles as bioisosteres. Lett. Drug Des. Discov. 2009;6:21–28. [Google Scholar]
- 8.Kremmydas G.F., Tampakaki A.P., Georgakopoulos D.G. Characterization of the biocontrol activity of Pseudomonas fluorescens strain X reveals novel genes regulated by glucose. PLoS One. 2013;8 doi: 10.1371/journal.pone.0061808. e61808. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Briones A.M., Okabe S., Umemiya Y., Ramsing N.B., Reichardt W., Okuyama H. Ammonia-oxidizing bacteria on root biofilms and their possible contribution to N use efficiency of different rice cultivars. Plant Soil. 2003;250:335–348. [Google Scholar]
- 10.Yu X., Ai C., Xin L., Zhou G. The siderophore-producing bacterium, Bacillus subtilis CAS15, has a biocontrol effect on Fusarium wilt and promotes the growth of pepper. Eur. J. Soil Biol. 2011;47:138–145. [Google Scholar]
- 11.Idris E.E., Iglesias D.J., Talon M., Borriss R. Tryptophan-dependent production of indole-3-acetic acid (IAA) affects level of plant growth promotion by Bacillus amyloliquefaciens FZB42. Mol. Plant Microbe Interact. 2007;20:619–626. doi: 10.1094/MPMI-20-6-0619. [DOI] [PubMed] [Google Scholar]
- 12.Saitou N., Nei M. The neighbor-joining method: a new method for reconstructing phylogenetic trees. Mol. Biol. Evol. 1987;4:406–425. doi: 10.1093/oxfordjournals.molbev.a040454. [DOI] [PubMed] [Google Scholar]
- 13.Felsenstein J. Evolutionary trees from DNA sequences: a maximum likelihood approach. J. Mol. Evol. 1981;17:368–376. doi: 10.1007/BF01734359. [DOI] [PubMed] [Google Scholar]
- 14.Fitch W.M. Toward defining the course of evolution: minimum change for a specific tree topology. Syst. Zool. 1971;20:406–416. [Google Scholar]
- 15.Felsenstein J. Confidence limits on phylogenies: an approach using the bootstrap. Evolution. 1985;39:783–791. doi: 10.1111/j.1558-5646.1985.tb00420.x. [DOI] [PubMed] [Google Scholar]
- 16.Chin C.S., Alexander D.H., Marks P., Klammer A.A., Drake J., Heiner C., Clum A., Copeland A., Huddleston J., Eichler E.E., Turner S.W., Korlach J. Nonhybrid, finished microbial genome assemblies from long-read SMRT sequencing data. Nat. Methods. 2013;10:563–569. doi: 10.1038/nmeth.2474. [DOI] [PubMed] [Google Scholar]
- 17.Jensen L.J., Julien P., Kuhn M., von Mering C., Muller J., Doerks T., Bork P. eggNOG: automated construction and annotation of orthologous groups of genes. Nucleic Acids Res. 2008;36:D250–D254. doi: 10.1093/nar/gkm796. [DOI] [PMC free article] [PubMed] [Google Scholar]