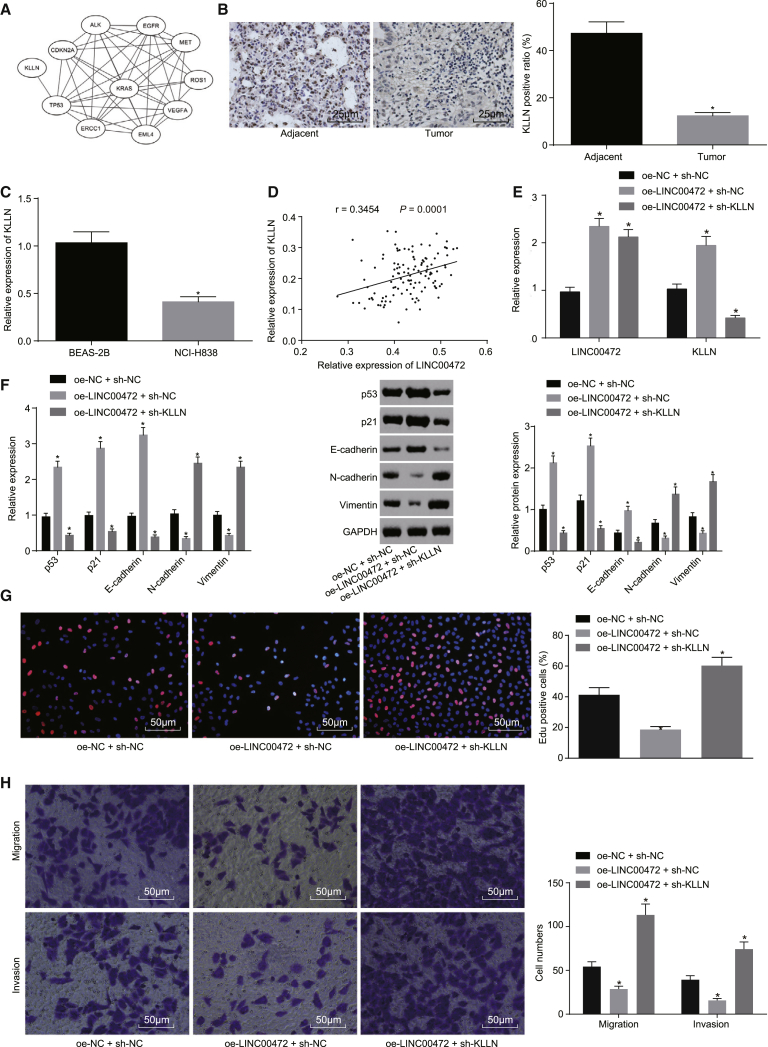
Figure 6.
Overexpressed LINC00472 Activates the p53-Signaling Pathway by Upregulating the Expression of KLLN to Suppress the EMT, Migration, and Invasion of NCI-H1299 Cells
NCI-H1299 cells were co-transfected with oe-LINC00472 and sh-NC or sh-KLLN, with oe-NC or sh-NC as the control. (A) Correlation analysis of core target genes and NSCLC-related known genes. Each ellipse in the figure represents a gene, and the line between the ellipses indicates the interaction between the two genes. (B) The immunohistochemistry showing the expression of KLLN in NSCLC tissues (the original magnification is ×400). *p < 0.05 versus the adjacent normal tissues. (C) The qRT-PCR showing the expression of KLLN in NSCLC cells, normalized by GAPDH. *p < 0.05 versus the BEAS-2B cells. (D) Correlation analysis of LINC00472 and KLLN expression. (E) The qRT-PCR showing the transfection efficiency of each group. (F) The qRT-PCR and western blot analysis showing the expression of p53 and epithelial marker (E-cadherin) and interstitial markers (N-cadherin and Vimentin) related to the EMT, normalized by GAPDH. (G) The EdU assay showing cell proliferation (the original magnification is ×200). (H) The Transwell assay showing cell migration and invasion (the original magnification is ×200). *p < 0.05 versus the oe-NC + sh-NC group (E–H). The representative measurement data were expressed as mean ± SD. Comparisons between the NSCLC tissues and adjacent normal tissues were analyzed by paired t test, and other comparisons between two groups were analyzed by unpaired t test. Comparisons among multiple groups were analyzed by one-way ANOVA. Pearson correlation analysis was performed between LINC00472 and KLLN. The experiment was repeated three times. LINC00472, long intergenic non-protein-coding RNA 472; NSCLC, non-small-cell lung cancer; EMT, epithelial-mesenchymal transition; GAPDH, glyceraldehyde-3-phosphate dehydrogenase.