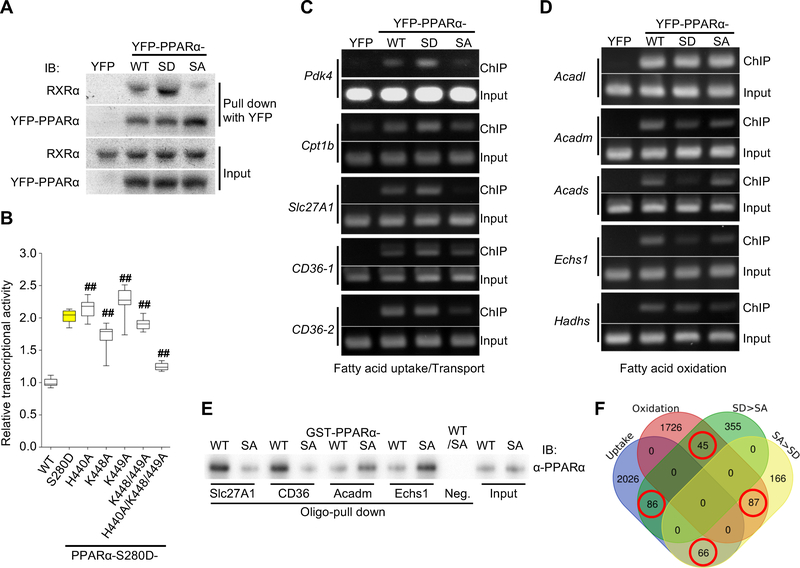
Figure 4. PPARα-Ser280 phosphorylation enhances both interaction with RXRα and PPRE binding.
(A) Representative immunoblots showing the interaction between YFP-PPARα-wild type (WT), -S280D (SD) or -S280A (SA) mutant and RXRα in cardiomyocytes (CMs) in vitro. YFP alone was used as a control. (B) PPRE-luciferase reporter assay using a series of alanine mutations to evaluate the effect of the indicated basic residues on the activity of PPARα-S280D (n = 12). ## p<0.001 compared to PPARα-S280D. (C) Chromatin immunoprecipitation (ChIP) assays using CMs transduced with adenovirus (Ad)-YFP-PPARα-WT, -SD, -SA, or YFP alone as a control. DNA was amplified by PCR with specific primers flanking the promoter of the indicated genes containing the PPARα-binding motif. PCR using input DNA as template served as an internal control. The data shown are representative of three independent experiments. (D) ChIP assays using specific primers flanking the promoter of the indicated genes containing the PPARα-binding motif. The data shown are representative of three independent experiments. (E) Double-stranded oligo pull-down assays, using biotinylated oligos containing the specific PPRE sequences in the indicated gene promoters. Recombinant GST-PPARα-WT or -SA was subjected to in vitro kinase assays using recombinant GSK-3α prior to the oligo pull-down assays. An oligo containing a PPRE with mutations in four base pairs was used as a negative control. (F) Venn diagram showing the number of genes in the rat genome containing the specific PPRE/DR1 motif (shown in Figure S5F) in their promoters. The numbers of overlapping genes in the Venn diagram (red circle) were significantly different (p=0.0002, Fisher’s exact test). See also Figure S5.