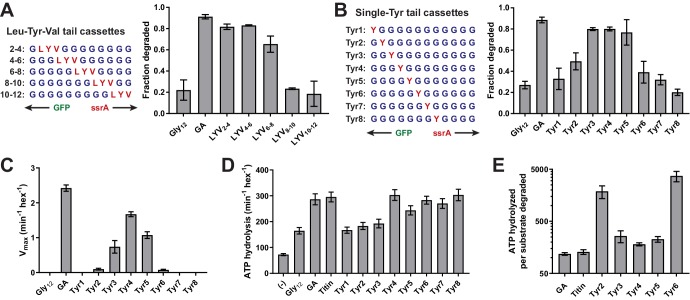
Figure 2. A small subset of tail residues mediate grip during GFP unfolding.
(A) Fraction intracellular degradation for substrates with tails containing LYV tripeptides in otherwise all-glycine cassettes. Gly12 and GA substrates were included as internal controls. (B) Fraction intracellular degradation for substrates with tails containing one tyrosine (Y) in otherwise all-glycine cassettes. Gly12 and GA substrates were included as internal controls. (C) Vmax values from Michaelis-Menten analysis of degradation of purified substrates with single-tyrosine cassettes. (D) Rates of ATP hydrolysis by ClpXΔN (0.1 μM hexamer) in the presence of ClpP (0.3 μM 14-mer) in the absence (–) or presence of different substrates (15 μM monomer). (E) ATP cost of degrading substrates with single-tyrosine cassettes. Note that the Y-axis is logarithmic. In all panels, values represent averages (± S.D.) of three biological replicates.