FIGURE 4.
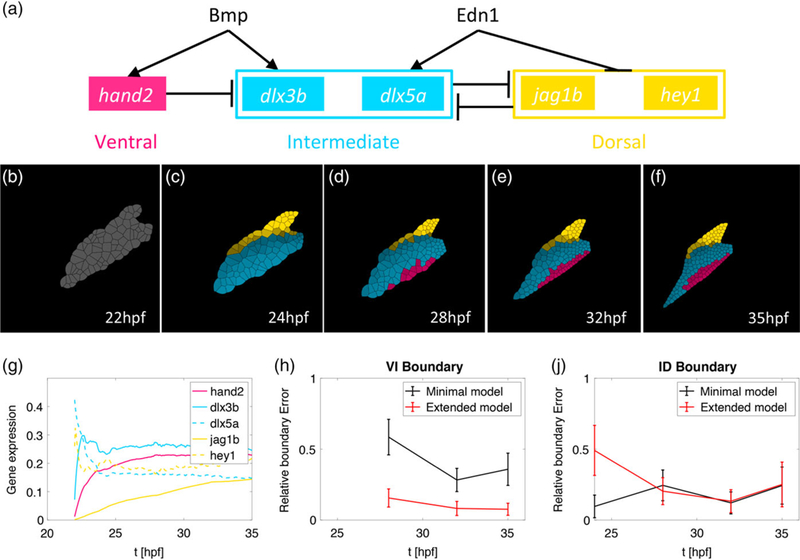
Computational modeling reveals that multiple genes with distinct temporal expression patterns stabilize V-I domain boundary formation. (a) The gene regulatory network (GRN) where individual genes specify the ventral (pink), intermediate (blue), and dorsal (yellow) identities. This model is an extension of the original model introduced in Meinecke et al., 2018 (b-f) simulation results of the spatial gene expression profiles incorporating measured temporal trajectories. The hey1 profile particularly drives dorsal domain establishment to earlier time points. (g) The time-dependent gene expression profiles in the simulation match those measured in vivo in zebrafish (Figure 3) for the time interval 22–35 hpf. (h, j) The y-axis depicts the normalized error of the boundary position in the computational simulations compared to the position observed by quantifying hand2 and dlx5 transgene expression in Meinecke et al., 2018. The solid line represents the mean value over 100 stochastic simulations and the error bars one standard deviation. (h) Comparison with the original model reveals that the parallel patterning of the three spatial domains with different genes with distinct temporal expression profiles stabilizes the accuracy of positioning the V-I boundary. (j) at the I-D boundary, initially higher error due to the early expression of the dorsal gene hey1 resolves to similar error levels at later time points
