Figure 5. BRAFV600E mutation enhances NHEJ by increasing XLF expression in thyroid cancer cells.
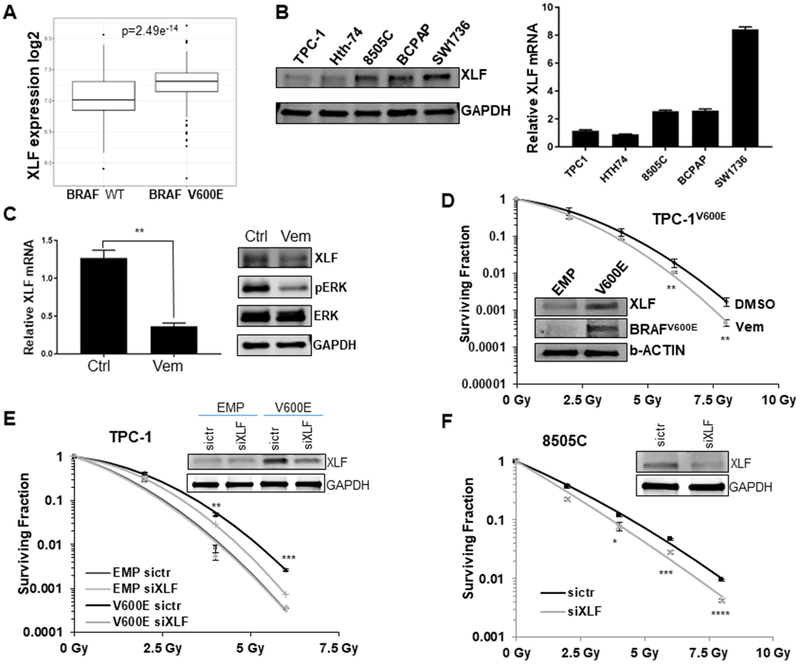
(A) Correlation of XLF expression with BRAFV600E mutation in TCGA thyroid cancer (THCA) data. Box plot showing the relationship between XLF expression and BRAFV600E mutation status. Expression was measured from primary tumor and is reported as log2 of the normalized count plus 1. A linear model shows a significant association (p=2.4e−14) of XLF expression with BRAFV600E mutation status in thyroid cancer. (B) A panel of thyroid cancer cell lines with BRAF wild type (U-Hth-74, TPC1) or V600E mutation (8505C, BCPAP, SW-1736) were analyzed for XLF protein by immunoblotting (left) and XLF mRNA levels by quantitative RT-PCR (right). (C) Exponentially growing BCPAP cells were treated with 100 nM vemurafenib or vehicle control (Ctrl) and analyzed for XLF mRNA levels by quantitative RT-PCR (left) and XLF protein by immunoblotting (right). **p<0.01 Vem vs. Ctrl. (D) TPC-1 cells expressing BRAF V600E (TPC-1 V600E) were cultured in media containing 100 nM vemurafenib (Vem) 3 hrs prior to irradiation with 0 (no IR), 2, 4, 6 and 8 Gy doses, followed by radiation clonogenic survival assays. Inset shows the upregulation of XLF and BRAFV600E in TPC-1 V600E cells by immunoblotting. **p<0.01 Vem vs. DMSO. BRAFV600E specific antibody confirms V600E expression. (E) Stable TPC-1 cells expressing BRAFV600E (TPC-1 V600E) or empty vector control (TPC-1 EMP) were transfected with siRNA of control (sictr) or XLF (siXLF) for 24 hrs. Then the cells were irradiated with 0 (no IR), 2, 4, 6 and 8 Gy doses, followed by radiation clonogenic survival assays. Inset shows the knockdown of XLF in TPC-1 V600E cells by immunoblotting with GAPDH as loading control. **p<0.01, ***p<0.001 V600E sictr vs. V600V siXLF. (F) BRAFV600E mutant 8505C cells were transfected with siRNA of control (sictr) or XLF (siXLF) for 24 hrs. Then the cells were irradiated with 0 (no IR), 2, 4, 6 and 8 Gy doses, followed by radiation clonogenic survival assays. Inset shows the knockdown of XLF in 8505C cells by immunoblotting with GAPDH as loading control. Each dose was prepared in triplicate per experiment, and no less than 2 experiments were performed. *p<0.05, ***p<0.001, ****p<0.0001
