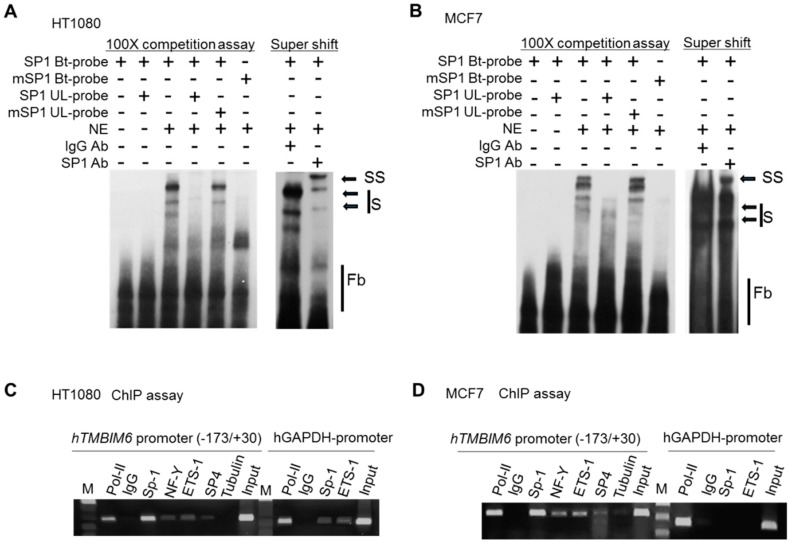
Figure 4.
EMSA and ChIP assay showing Sp1 binding to the human TMBIM6 promoter. (A,B) Nuclear extract (NE) was prepared from HT1080 (A) and MCF7 (B) cells. Double-stranded 35-bp oligonucleotide probe specific to Sp1 proximal site (−25/+7) was generated and labeled with biotin. The labeled Sp1-p probe was incubated alone (lane 1), with unlabeled probe (lane 2), with the NE (lane 3), with the NE and 200-fold excess amount of unlabeled homologous probe (lane 4), with 200-fold excess mutant probe (lane 5), and with mutant-labeled probe that failed to form a complex with the NE (lane 6). DNA–protein complexes that formed are indicated (S), unbound probes indicated as free probes (Fb). In lanes 7–9, the super shift assay was performed by pre-incubating the NE with probe for 20 mins on ice before adding the specific antibody. Then, the specific probe was added and further incubated for 20 mins on ice, with normal IgG as the negative control in lane 7 and anti-Sp1 monoclonal antibody in lanes 8, respectively. The super shift is indicated as SS. (C,D) ChIP was carried out with HT1080 (C) and MCF7 (D) cells; antibodies for RNA polymerase II (pol-II), Sp1, ETS-1, NF-Y, and SP4 were used, with tubulin and IgG as the negative control. Immunoprecipitated chromatin was amplified by PCR with primers specific to region −173 to +30 of the TMBIM6 gene. The GAPDH locus was used as the control. M: DNA marker.