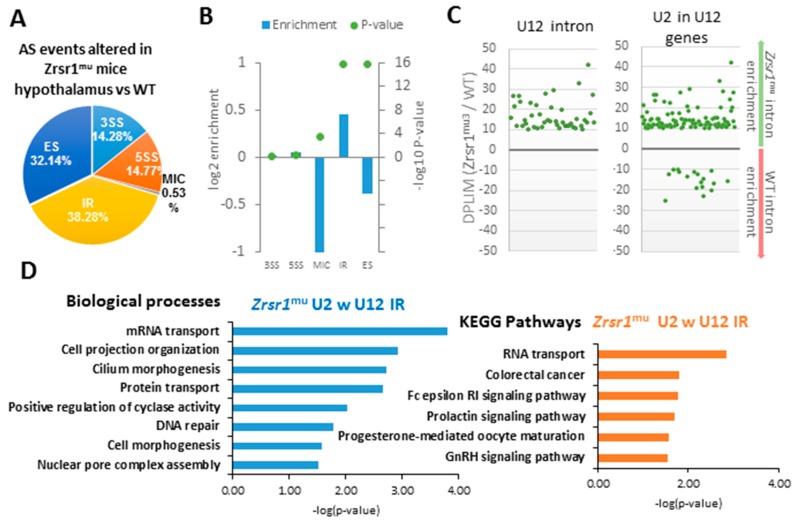
Figure 2.
RNA-seq analysis of differential alternative splicing (AS) in Zrsr1mu mouse hypothalamus. (A) Distribution of the categories of AS events detected in the analysis. 3SS, alternative 3′ splice sites; A5SS, alternative 5′ splice sites; ES, exon skipping; IR, intron retention; MIC, alternative microexon. (B) Enrichment of the different categories of AS in our analysis in relation with the total number of events present in the vast-tools database. (C) Distribution of differentially retained introns that correspond to U12 introns (left) and U2 introns located in a U12-intron-containing gene (right) detected in our analysis. (D) GO terms associated to biological processes and KEGG pathways enriched in the U12-containing genes detected as differentially spliced.