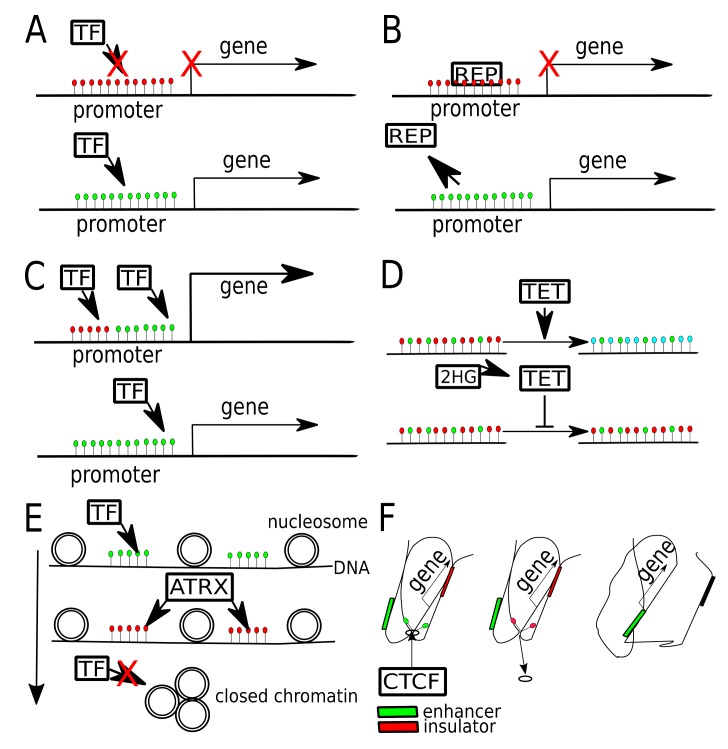
Figure 1.
Different ways that DNA methylation may modulate gene expression in gliomas. (A) Gene promoter hypermethylation (red dots—5mC) may cause gene expression silencing, by blocking transcription factor (TF) binding. Once promoter gets unmethylated (green dots—unmethylated C), gene expression may occur. (B) Gene promoter, when hypermethylated binds transcriptional repressor (REP). Once promoter gets unmethylated, repressor is released, allowing gene expression. (C) Binding of two TFs to one promoter boosts gene expression, one of TFs binds preferentially to methylated, while the other to unmethylated DNA. (D) In physiological conditions TET proteins oxidize 5mC (red dots) to 5hmC (blue dots) and later to other derivatives (5fC, 5caC). When TET gets inhibited by 2HG (2-hydroxyglutarate), produced by mutant IDH1/2, it fails to demethylate DNA, thus maintaining global DNA hypermethylation. (E) DNA methylation is related with chromatin openness, due to chromatin chaperons, that are sensitive to DNA and histone being activated or repressed. ATRX protein may bind to methylated DNA and lead to heterochromatin formation, blocking access of TFs. (F) Hypermethylation of CTCF binding site causes CTCF unbinding and change of chromatin conformation, which leads to exchange of insulator by enhancer in a close proximity of a gene.