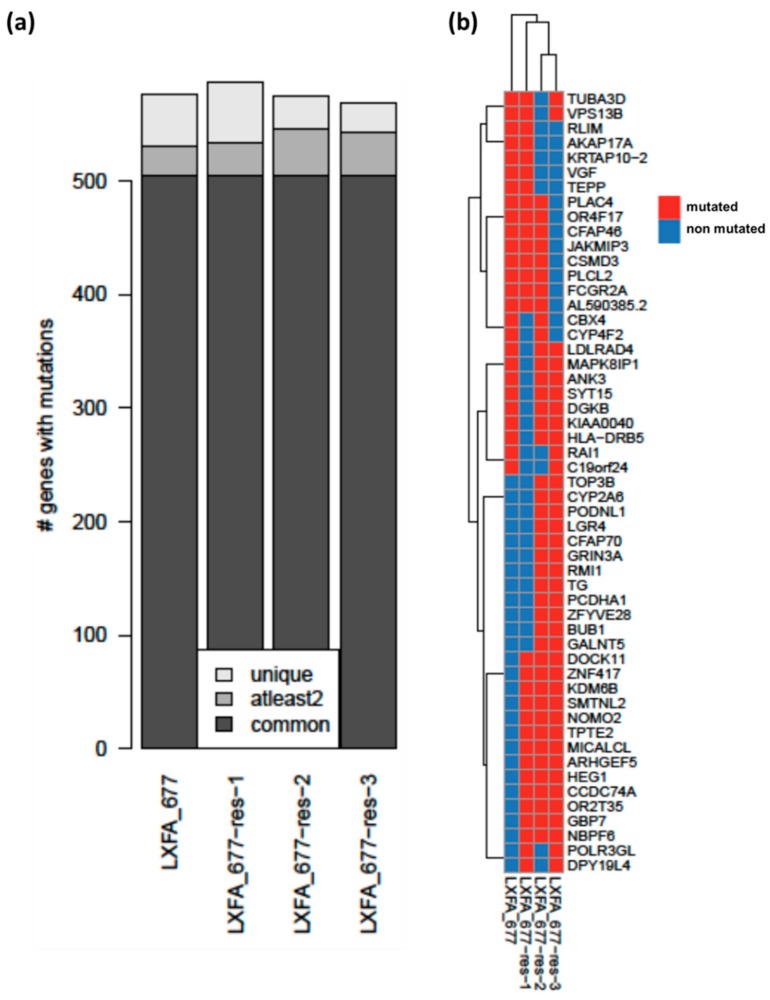
Figure 3.
Results of whole exome sequencing analysis of four different LXFA 677 sublines. (a): Mutational landscape of the four LXFA 677 sublines with more than 85% (n = 504) of genes with mutations being shared across clones. (b): Hierarchical clustering of genes with differing (=at least two; n = 53) mutation status. The whole exome sequencing was performed using the variant effect predictor (VEP). Candidate mutations were annotated and filtered considering only variants with moderate or high protein impact and those being rare in healthy populations (<1% in gnomAD). Mutations shared by all three resistant clones were annotated with protein functions using SIFT and Polyphen predictions from SNPnexus as depicted in detail in Table S2.