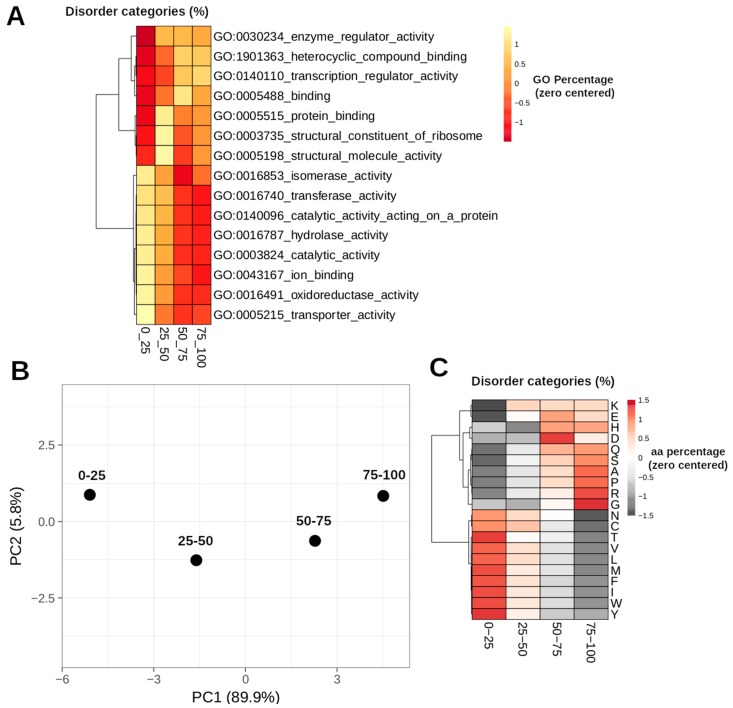
Figure 2.
General characteristics of the proteins according to the disorder content of each category defined in this work. (A) Heatmap of the enriched Gene Ontology (GO) terms between the four defined categories for a p-value < 0.01. Colors represent the percentage of each GO category with scaling applied to rows and the values centered to zero. (B) Principal Component Analysis (PCA) analysis of the aa composition of the proteins that constitute each category. Unit variance scaling is applied to relative aa composition of the proteins of each category; Singular Value Decomposition (SVD) with imputation is used to calculate principal components. X and Y axis show principal component 1 and principal component 2 which explain 89.9% and 5.8% of the total variance, respectively. (C) Heatmap of the aa composition of the different categories of protein disorder. The rows were centered, and scaling was applied. The rows were then clustered using correlation distance and complete linkage.