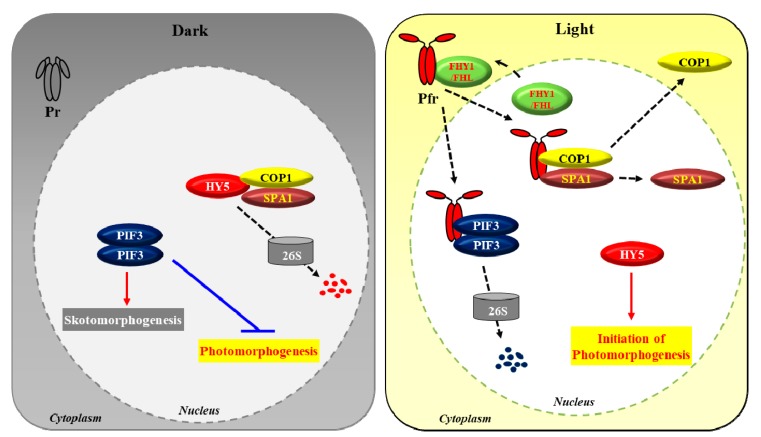
Figure 2.
A simplified view of the phytochrome-mediated photomorphogenesis in A. thaliana. For simplicity, we use PIF3 as a representative of PIFs, HY5 as a representative of the master transcription factors of photomorphogenesis, and SPA1 as a representative of the SPA proteins. In the dark (left panel), phytochromes are biosynthesized as the inactive Pr forms, staying in the cytoplasm. Meanwhile, PIF3 proteins accumulate in the nucleus and regulate the expression of genes to prevent photomorphogenesis (shown as a blue T bar) by promoting skotomorphogenesis (shown as a red arrow). In addition, the COP1-SPA1 complex constantly degrade the expressed HY5 proteins via the ubiquitin 26S proteasome pathway to prevent photomorphogenesis (shown as a dotted black arrow). In the light (right panel), the photoactivated Pfr forms of phytochromes translocate into the nucleus (in the case of phyA, FHY1 and FHL are the facilitators for the import), where they can interact with downstream signaling components. They inactivate PIF3 by inducing protein degradation and the COP1-SPA1 complex by inducing its dissociation and subsequent nuclear exclusion of COP1, which all contribute to the accumulation of the master transcription factors for photomorphogenesis, such as HY5. Finally, HY5 induces the expression of light-responsive genes for photomorphogenic development. Movements of proteins are also shown as dotted arrows.