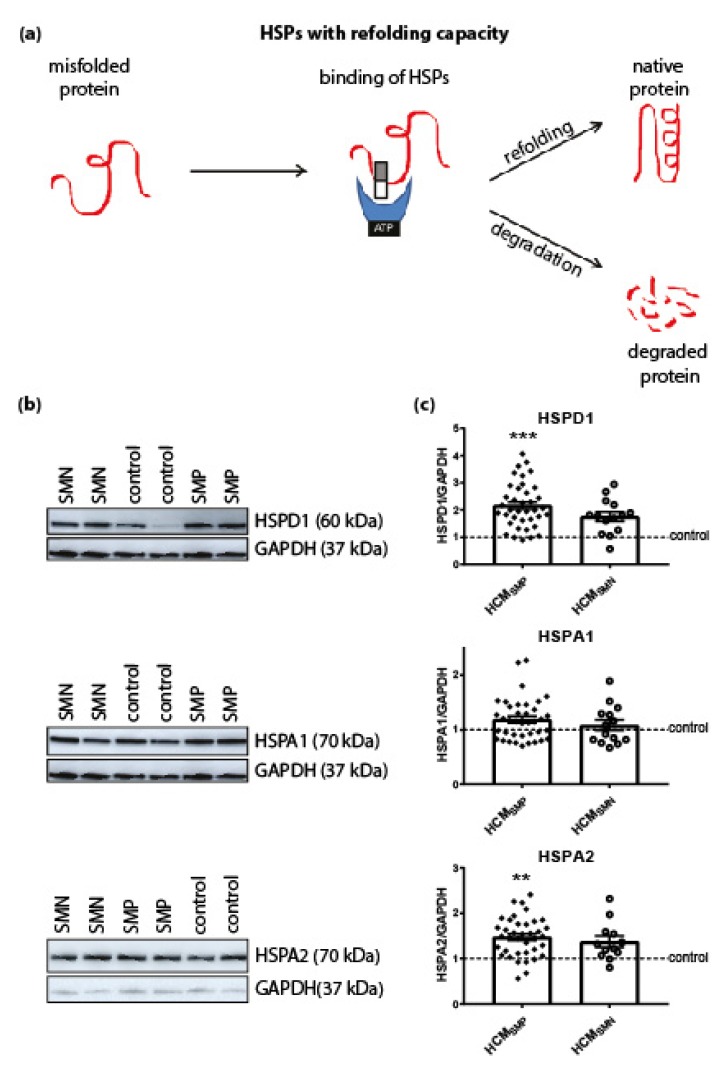
Figure 2.
HSPs with a refolding capacity. (a) HSPs with ATPase activity and stabilizing HSPs bind to the misfolded protein to stabilize and further process it. HSPs with a refolding capacity either refold the misfolded protein to its native structure or, if refolding is impossible, the HSPs assist in the degradation pathways to degrade the misfolded protein. Figure reproduced and adapted with permission from Dorsch, L.M. et al., Pflügers Archiv—European Journal of Physiology, published by Springer Berlin Heidelberg, 2018. (b) Representative blot images for HSPD1, HSPA1, and HSPA2 expression. (c) Higher levels of HSPD1 and HSPA2 in HCMSMP (n = 38) compared to controls. HSPs with refolding capacity were unaltered in HCMSMN (HSPD1, HSPA1: n = 14; HSPA2: n = 12) compared to controls. There were no significant differences between HCMSMP and HCMSMN. Each dot in the scatter plots represents an individual sample. ** p < 0.01 and *** p < 0.001 versus controls.