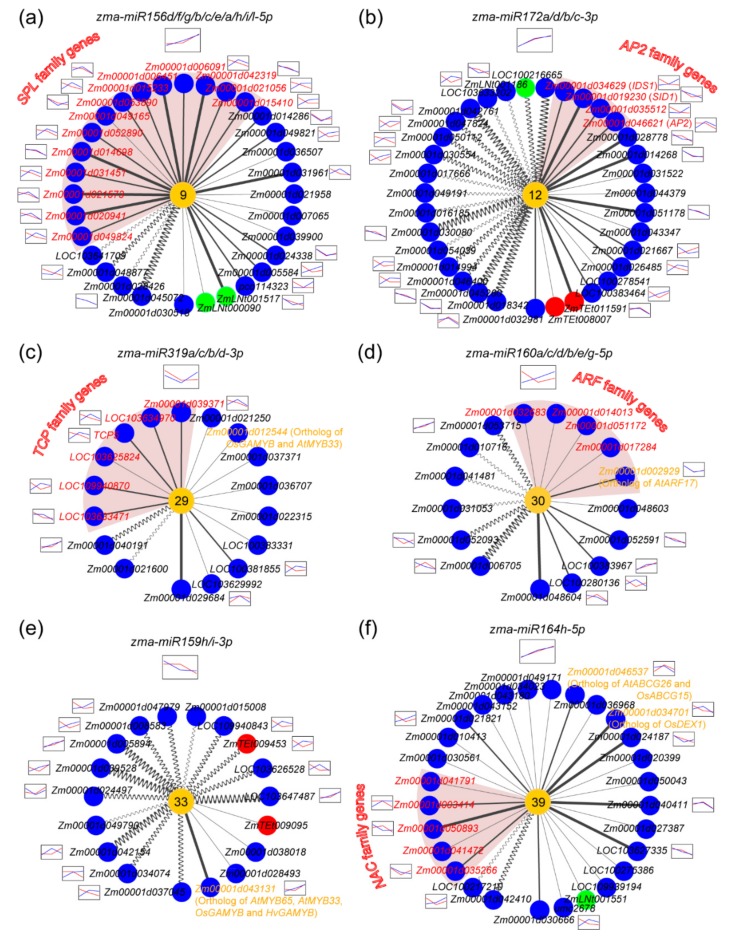
Figure 6.
The six known miRNA-mediated regulatory networks functionally associated with maize anther development. The six networks mediated by the known miRNAs of (a) zma-miR156, (b) zma-miR172, (c) zma-miR319, (d) zma-miR160, (e) zma-miR159, and (f) zma-miR164 are shown. The transcription expression patterns of corresponding miRNAs are shown at the top of the networks based on small RNA transcriptomes in maize lines L7 (marked in red curve) and L30 (marked in blue curve). The transcription expression patterns of ceRNA or target genes that were negatively associated with miRNA expression in at least one line are shown beside them. The names and the backgrounds of target genes in the same gene family in each network are marked in red. The names of genes with orthologs that participate in anther development in other plant species are shown in yellow. Other legend of this figure is the same as that of Figure 4 and Figure 5.