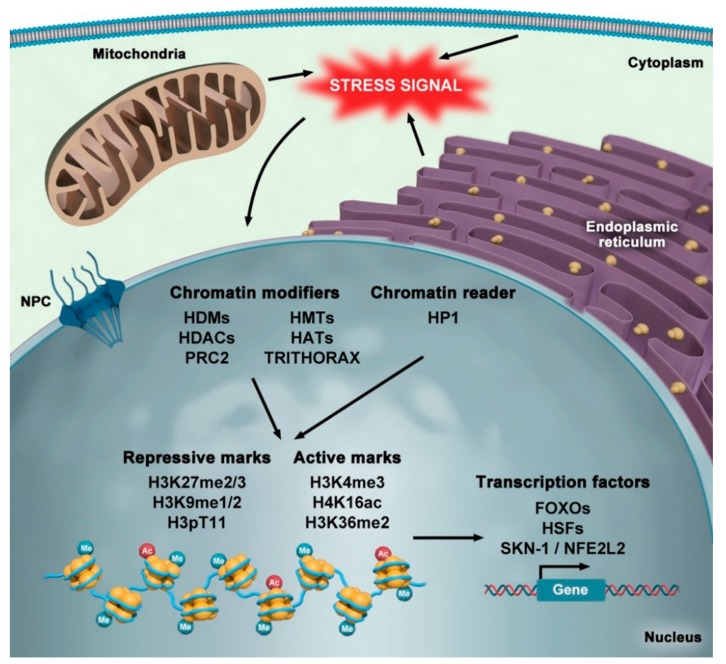
Figure 2.
Epigenetic responses to stress. Stress signals from outside the cell or from specific organelles, such as mitochondria or the endoplasmic reticulum, are detected and processed by different signaling pathways [146]. The transduction of these stimuli results in the dynamic regulation of epigenetic mechanisms, mediated by a broad variety of chromatin protein modifiers acting as intermediates to make an appropriate cellular response [147]. Chromatin modifiers can add or remove specific histone modifications, such as methyl, acetyl, or phosphate groups. These different marks alter binding of chromatin readers (e.g., HP1) as well as gene expression through the action of different transcription factors (e.g., FOXOs, HSFs, SKN-1/NFE2L2) [121,123,148,149,150,151,152]. A non-exhaustive list of chromatin modifiers and readers associated to stress are depicted (abbreviations: HATs, histone acetyl transferases; HDACs, histone deacetylases; HDMs, histone demethylases; HMTs, histone methyltransferases; HP1, heterochromatin protein 1; NPC, nuclear pore complex; PRC2, polycomb repressive complex 2).