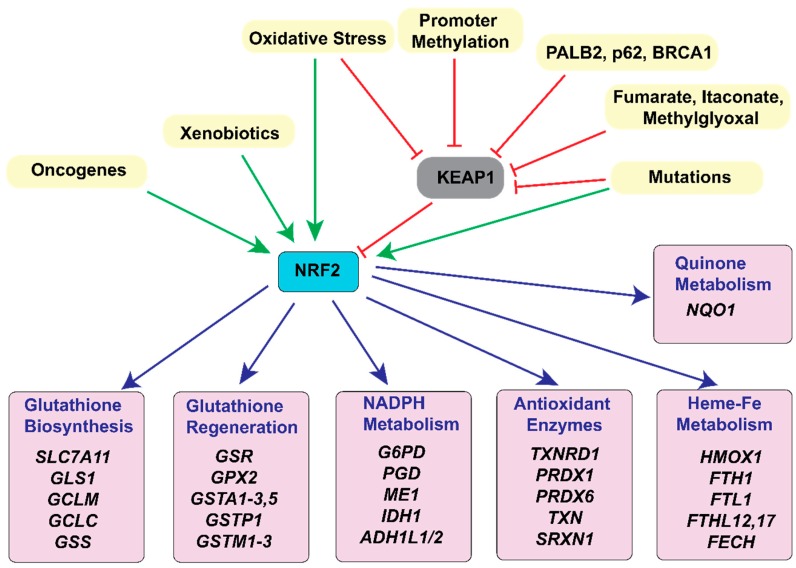
Figure 3.
KEAP1-NRF2 in the regulation of metabolism and cellular antioxidant response. Nuclear factor erythroid 2-related factor 2 (NRF2) is the regulator of antioxidant response in cancer. NRF2 is activated by gene mutations, oncogene activation, xenobiotics, or oxidative stress. Oxidative stress also inactivates the principal NRF2 inhibitor in cells, Kelch-Like ECH-associated protein 1 (KEAP1), by cysteine oxidation. KEAP1 is also inhibited by mutations, promoter methylation, binding with KEAP1-sequestering proteins like PALB2, BRCA1, and p62, and interaction with metabolites such as fumarate, itaconate, and methylglyoxal. Stabilized NRF2 regulates expression of several genes involved in redox homeostasis. Shown here are NRF2-target genes involved directly in redox balance. First are genes involved in glutathione (GSH) biosynthesis: solute carrier family 7 member 11 (SLC7A11), glutaminase1 (GLS1), glutamate-cysteine ligase, modifier subunit (GCLM), glutamate-cysteine ligase catalytic subunit (GCLC), and glutathione synthetase (GSS). Glutamate generated by GLS1, a NRF2 target gene, can also be utilized for GSH biosynthesis. The second set has genes involved in GSH recycling: glutathione-disulfide reductase (GSR), glutathione peroxidase 2 (GPX2), glutathione S-transferase Alpha (GSTA), glutathione S-transferase Pi (GSTP), and glutathione S-transferase Mu (GSTM). Third set has enzymes mediating NADPH metabolism: glucose-6-phosphate dehydrogenase (G6PD), phosphogluconate dehydrogenase (PGD), malic enzyme (ME1), isocitrate dehydrogenase (IDH1), aldehyde dehydrogenase 1 family, and member L1 and L2 (ADH1L1/2). Other metabolic enzymes shown in the 4th set are: thioredoxin 1 (TXN1), thioredoxin reductase 1 (TXNRD1), peroxiredoxin 1 (PRDX1), peroxiredoxin 6 (PRDX6) and sulfiredoxin 1 (SRXN1). Fifth/Sixth set are enzymes in Heme, Iron (Fe) and quinone metabolism: heme oxygenase 1 (HMOX1), ferritin heavy chain 1 (FTH1), ferritin light chain 1 (FTL1), ferritin heavy chain 1 pseudogene 12 (FTHL12), ferritin heavy chain 1 pseudogene 17 (FTHL17), ferrochelatase (FECH) and NAD(P)H quinone dehydrogenase 1 (NQO1). (Green arrows indicate direct and red arrows indirect mechanisms of NRF2 activation. Blue arrows direct to genes transcriptionally regulated by NRF2).