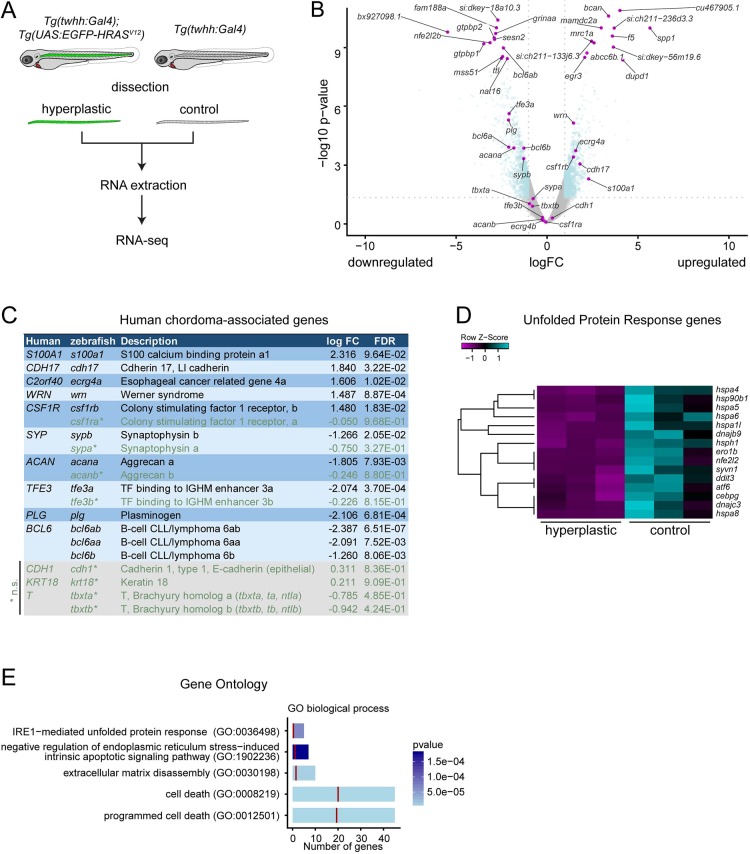
Fig. 5.
HRASV12-induced zebrafish chordomas have a deregulated UPR and suppress apoptosis. (A) Workflow of control and transformed notochord isolation. Notochords were dissected from twhh:Gal4 (wild-type morphology and transgenic base line) and twhh:Gal4;UAS:EGFP-HRASV12-expressing larvae at 8 dpf; see Materials and Methods for details. (B) Volcano plot depicting overall distribution of deregulated genes: gray, genes for which P<0.05; blue, genes with significant deregulation between control versus transformed notochords. Note that the zebrafish brachyury genes tbxta and tbxtb are unchanged but expressed. (C) Expression of human chordoma-associated genes in HRASV12-transformed zebrafish notochords; see text for details. Zebrafish orthologs of human chordoma genes are shown with all their orthologs: orthologs with changes considered to be not significant (n.s., green) are marked with asterisks, genes with no significantly altered ortholog are shown in gray boxes at the bottom. (D) Significantly deregulated UPR genes as per IPA pathway analysis of hyperplastic versus control zebrafish notochords. (E) Gene ontology (GO) enrichment in hyperplastic zebrafish notochords also highlights deregulation of the UPR, ER stress, ECM dynamics and cell death. The vertical red bars represent the expected number of genes per GO term under random selection based on the reference list, Homo sapiens REFLIST (21,042 genes in total). See also Fig. S5.