Figure 2: Modeling and construction of Hsd17b4 mutants.
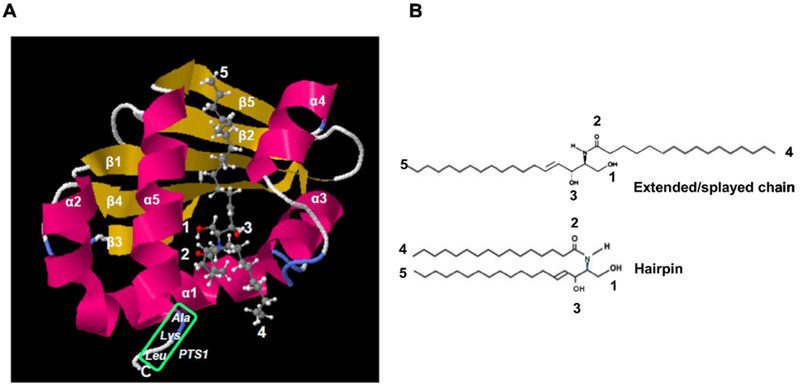
A. Molecular modeling of the Scp2-like domain using SwissDock with pdb file 1IKT (Scp2 like) and C16:0 ceramide (Zinc file 40164304.mol2) showing conformation with sphingosyl residue binding to protein (ΔG0’ = 10.3 kcal/mol, KD’ = 30 nM) and fatty acid residue accessible for membrane anchoring or immobilization on affinity gel as shown in Fig. 1A. Green box shows PTS1 sequence. Arrows point at residues indicated in legend at the upper left of image panel and B. B. Ext ended/splayed chain and hairpin conformation of C16:0 ceramide. The extended conformation is compatible with binding of ceramide shown in A.
