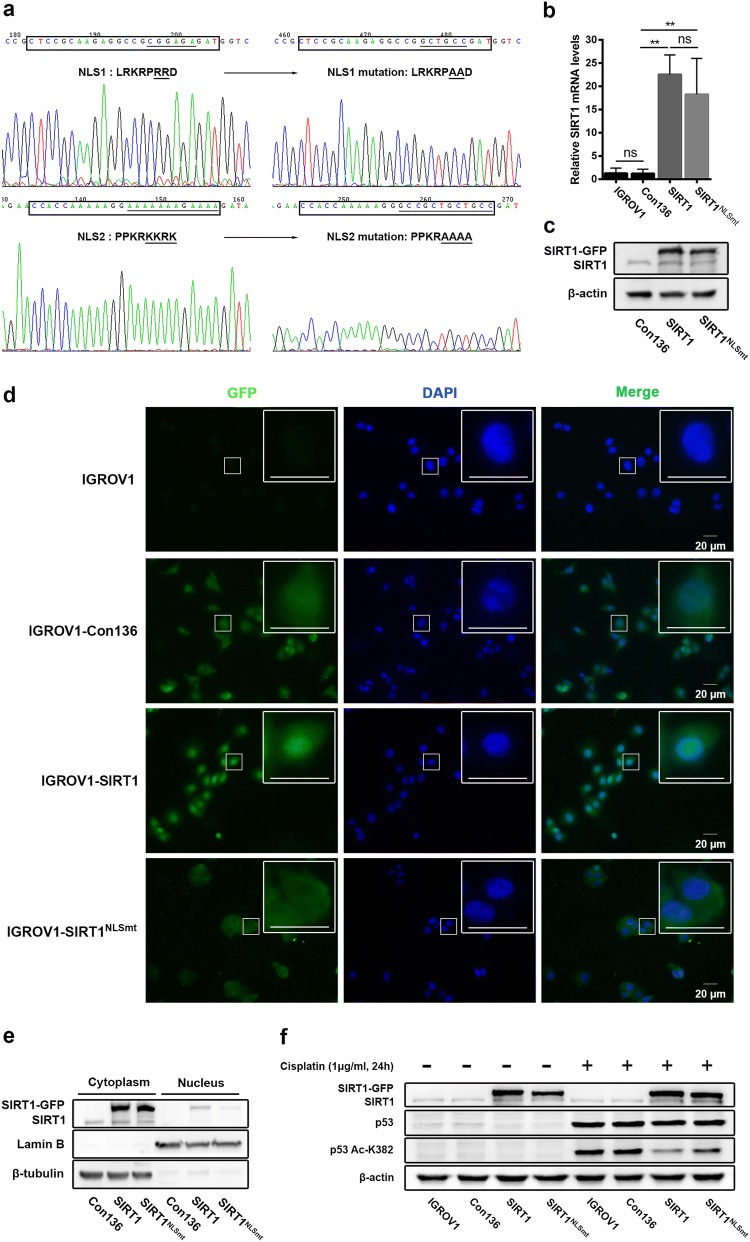
Fig. 1.
Analyses of SIRT1 subcellular location and expression in ovarian carcinoma IGROV1 cells overexpressing SIRT1 (SIRT1 cells) or NLS-mutated SIRT1 (SIRT1NLSmt cells). a The NLS sequence located at amino acids 33–40 (LRKRPRRD) (CTCCGCAAGAGGCCGCGGAGAGAT) was mutated to LRKRPAAD (CTCCGCAAGAGGCCGGCTGCCGAT). In addition, the other NLS sequence located at amino acids 231–238 (PPKRKKRK) (CCACCAAAAAGGAAAAAAAGAAAA) was mutated to PPKRAAAA (CCACCAAAAAGGGCCGCTGCTGCC). b SIRT1 mRNA levels as measured by real-time PCR in the parental (IGROV1), Con136, SIRT1 and SIRT1NLSmt cells. Each bar represents the mean ± SD (n = 3; **P < 0.01; ns, not significant). c Western blot analyses of SIRT1 expression in Con136, SIRT1 and SIRT1NLSmt cells. β-Actin was used as a loading control. d Immunofluorescence staining of GFP in parental (IGROV1), Con136, SIRT1 and SIRT1NLSmt cells (original magnification: × 400; scale bars: 20 μm). Boxes show the cell details at a higher magnification (scale bars: 20 μm). e SIRT1 expression in cytoplasmic and nuclear extracts from Con136, SIRT1 and SIRT1NLSmt cells. β-tubulin and lamin B were used as cytoplasmic and nuclear loading controls, respectively. f Protein levels of SIRT1, p53 and acetylated (K382) p53 before and after treatment with 1 μg/ml for 24 h in Con136, SIRT1 and SIRT1NLSmt cells as analyzed by western blotting. β-Actin was used as a loading control