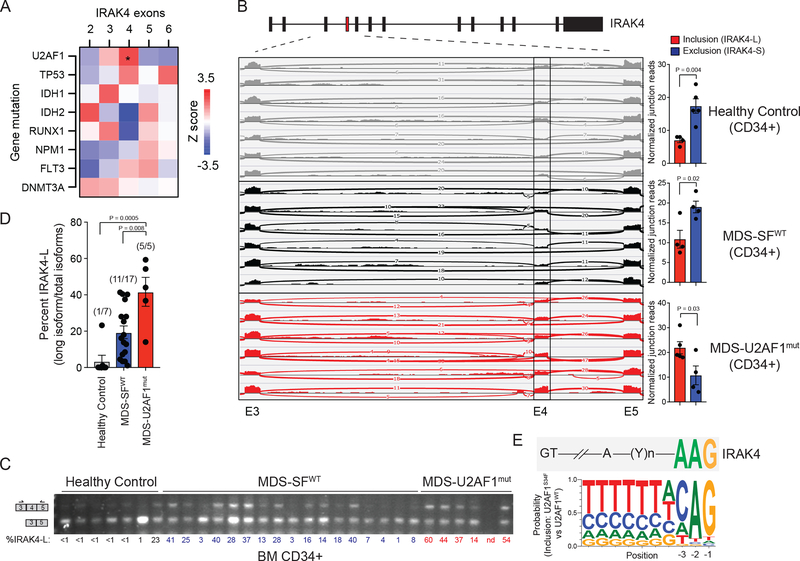
Figure 4. IRAK4-L expression is associated with U2AF1 mutations in MDS and AML.
(A) Heatmap was generated based on the Z-score correlation of individual exon’s expression to all somatic mutations. Each indicated mutation was regressed against the relative expression (after adjusting for total IRAK4 expression) of each IRAK4 exon (RPKM) in a multiple linear regression model. Shown are z-scores of partial correlations. A z-score of greater than |1.96| corresponds to P < 0.05. Six out of 160 AML patients exhibit U2AF1 mutations (Z = 2.43; *. P = 0.01). (B) Sashimi plots representing IRAK4 exon 4 inclusion or exclusion in healthy CD34+ cells (n = 7 samples), CD34+ cells isolated from MDS patients with no splicing factor (SFWT) mutations (MDS-SFWT; n = 6 samples), and from patients with mutation (mut) in U2AF1 (MDS-U2AF1mut; n = 6 samples) based on RNA-sequencing junction reads. Quantification of the junction reads for exon 4 are shown for healthy controls (n = 7), MDS-SFWT (n = 6), and MDS-U2AF1mut (n = 6). Exon 4 inclusion and exclusion reads were normalized to the total number of reads in each sample and plotted as a ratio (inclusion/total and exclusion/total). Two-sided t-tests were used for statistical analyses. (C) RT- PCR analysis of healthy CD34+ cells (n = 7 samples), CD34+ cells isolated from MDS patients with no splicing factor mutations (MDS-SFWT; n = 17 samples), and from patients with mutation in U2AF1 (MDS-U2AF1mut; n = 5 samples) using primers flanking IRAK4 exon 4. Densitometric quantification of IRAK4 exon 4 inclusion calculated as the ratio of the long isoform versus both isoforms is shown below each sample. (D) Densitometric quantification of IRAK4 exon 4 inclusion is summarized from panel (C). The number in parentheses indicates the number of samples exhibiting >10% expression of IRAK4-L relative to all IRAK4 isoforms. Two-sided t-tests were used for statistical analyses. (E) Consensus sequence motifs identified at the distal 3’ splice sites of exon exclusion events in U2AF1-S34F cells as compared to U2AF1 using publicly available data and visualized with weblogo software. Above the U2AF1-S34F consensus motifs is the sequence of the distal 3’ splice site of IRAK4 exon 4. All data represent the mean ± s.e.m.