Figure 5.
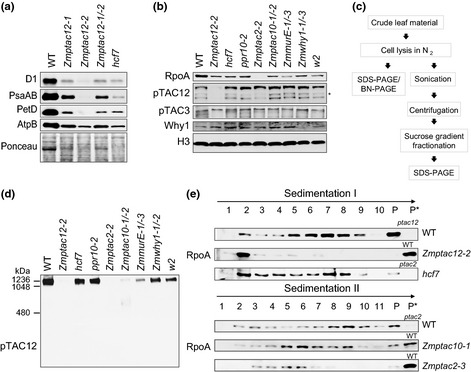
PEP‐complex assemblies in wild‐type (WT) and chlorophyll‐deficient maize mutants. (a) Immunoblot analyses of photosynthetic complex subunits. Labels on the left indicate the antibodies utilized for immunoblotting. Ponceau staining is shown to demonstrate equal loading of protein samples (25 μg) and abundance of RbcL (large subunit of Rubisco). (b) Accumulation of nucleoid‐associated proteins in chlorophyll‐deficient mutants (designation given above). Total proteins (25 μg) were separated by SDS‐PAGE and immunoblotted using antibodies against RpoA, pTAC12, pTAC3 and Why1. Histon H3 was used as loading control. Asterisk marks a band not detected in other immunoblots; its significance is not clear. (c) Flow chart of experimental procedure. (d) Separation of protein complexes from chlorophyll‐deficient mutants by BN‐PAGE and subsequent analyses of PEP‐complex assembly by immunoblotting with the anti‐pTAC12 antibody. Approximately 50 μg of total leaf protein (same preparations as above) from the base section of second leaves were loaded per lane. The gel was run in parallel with the SDS‐gel (shown in panel b) to ensure equal loading. (e) Sucrose‐gradient sedimentation of total leaf extracts from the basal half of the second leaf. Proteins from WT and indicated mutants were solubilized in Triton X‐100 protein lysis buffer with sonication, and soluble fractions were run through sucrose gradients. Fractions were collected and immunoblotted with an antibody against RpoA. Sedimentation I and II indicate two independent experiments. Proteins used in this experiments were isolated from 7‐ to 10‐d‐old maize seedlings. P, pelleted material; P*, pelleted material from WT or mutant leaf samples, respectively.
