Figure 2.
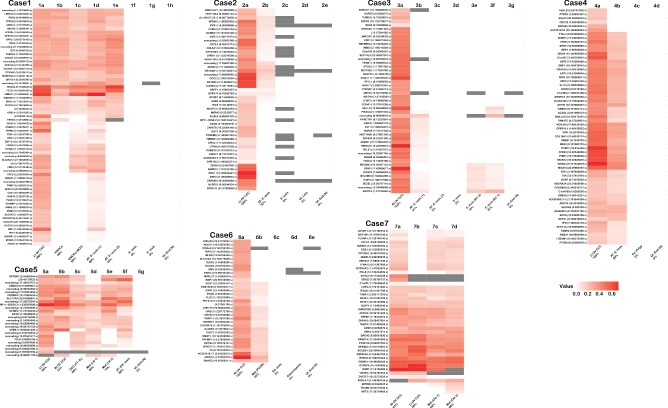
Heat map summary of verified somatic SNVs (s) and INDELs (i) in primary CCC tumours and other related or benign specimens from each case. Whenever possible, multiple samples of tumour, metastatic disease, putative precursor (including endometriosis) and/or benign lesion were examined from each case (see also supplementary material, Table S1, Figure S4). In all cases the left‐most specimen is the primary tumour used in WGSS and samples are ordered using their presumed relationship to the primary tumour, from other ovarian sites, metastatic sites, endometriosis (with and without atypia) to benign lesions and normal uterine endometrium. Patient‐matched germline DNA was used as the reference in both WGSS and deep sequencing confirmation experiments; intensity of red increases with allelic frequency; grey bars denote positions with insufficient coverage. Sub‐specimens are labelled under each panel; the percentage listed in the label refers to the conservation of somatic mutations compared to the index tumour specimen (see also supplementary material, Figure S3). Gene symbol, genomic position and mutation type is noted at the left of each panel; additional detail can be found in Tables S1 and S2 (see supplementary material): Lt OV, left ovary; Rt OV, right ovary; E‐osis, endometriosis; AT‐E‐osis, atypical endometriosis; ES‐osis, endosalpingiosis; Met, metastasis; EN‐Polyp, endometrial polyp; Ut‐End(N), uterine endometrium (normal); FT‐IL, Fallopian tube intraluminal fragment; LN, lymph node; BOT, borderline ovarian tumour; Ut‐Leio, uterine leiomyoma; RC, rectosigmoid colon; Om, omentum; PCDS, posterior cul‐de‐sac
