Summary
The ubiquitin‐like modifier (UBL) SUMO (Small Ubiquitin‐Like Modifier) regulates protein function. Structural rather than sequence homology typifies UBL families. However, individual UBL types, such as SUMO, show remarkable sequence conservation. Selection pressure also operates at the SUMO gene copy number, as increased SUMO levels activate immunity and alter flowering time in Arabidopsis.
We show how, despite this selection pressure, the SUMO family has diversified into eight paralogues in Arabidopsis. Relationships between the paralogues were investigated using genome collinearity and gene tree analysis. We show that palaeopolyploidy followed by tandem duplications allowed expansion and then diversification of the SUMO genes.
For example, Arabidopsis SUMO5 evolved from the pan‐eudicot palaeohexaploidy event (gamma), which yielded three SUMO copies. Two gamma copies were preserved as archetype SUMOs, suggesting subfunctionalization, whereas the third copy served as a hotspot for SUMO diversification.
The Brassicaceae‐specific alpha duplication then caused the duplication of one archetype gamma copy, which, by subfunctionalization, allowed the retention of both SUMO1 and SUMO2. The other archetype gamma copy was simultaneously pseudogenized (SUMO4/6). A tandem duplication of SUMO2 subsequently yielded SUMO3 in the Brassicaceae crown group. SUMO3 potentially neofunctionalized in Arabidopsis, but it is lost in many Brassicaceae. Our advanced methodology allows the study of the birth and fixation of other paralogues in plants.
Keywords: evolution, immunity, neofunctionalization, palaeoploidy, paralogue, protein modification, SUMO, ubiquitin‐like modifier
Introduction
Post‐translational modifications (PTMs) set a reversible mark on proteins, altering their function (van der Veen & Ploegh, 2012). The first polypeptide that was discovered to act as a PTM was ubiquitin (Ub), a highly conserved 76‐residue polypeptide. Ub and ubiquitin‐like modifiers (UBLs) are typified by their β‐grasp fold, which generates a highly stable tertiary structure resistant to environmental perturbations, such as heat (Burroughs et al., 2012; Vierstra, 2012; Callis, 2014). There is limited sequence identity between UBL types, yet remarkable sequence conservation is seen for individual UBL types across eukaryotes. For example, Ub is 96% identical between plants, yeast and mammals (Vierstra, 2003).
A prominent UBL type is the Small Ubiquitin‐Like Modifier (SUMO), which is conserved across eukaryotes (Miura & Hasegawa, 2010; Flotho & Melchior, 2013; Jentsch & Psakhye, 2013). Its conjugation is primarily associated with nuclear processes, such as nucleocytoplasmic transport, gene regulation, chromatin remodelling, DNA repair and DNA replication (Miller et al., 2010b, 2013; Flotho & Melchior, 2013). SUMO is translated as a precursor that undergoes C‐terminal processing by SUMO proteases (also known as ubiquitin‐like proteases or ULPs). The processing exposes a C‐terminal diglycine (diGly) motif essential for conjugation. Mature SUMO is conjugated to substrates via the E1 SUMO Activating Enzyme dimer (SAE1/2) and the E2 SUMO Conjugating Enzyme (SCE1) (Saracco et al., 2007; Castano‐Miquel et al., 2013). On conjugation (SUMOylation), an isopeptide bond is formed between the carboxyl terminus of mature SUMO and the acceptor lysine (Lys) side chain. SUMOylation is an essential process, with mutations causing embryonic lethality in mice and the model plant Arabidopsis (Arabidopsis thaliana) (Saracco et al., 2007; Wang et al., 2014). E3 ligases can promote SUMOylation (Flotho & Melchior, 2013). In Arabidopsis, two E3 ligases have been characterized. Loss of the E3 ligase SIZ1 (SAP AND MIZ 1) causes dwarfism, early flowering, altered responses to abiotic stresses and activation of plant immunity (Miura & Hasegawa, 2010; Park et al., 2011). By contrast, the E3 ligase High Ploidy2 (HPY2/MMS21) represses endocycle onset in meristems (Huang et al., 2009; Ishida et al., 2009, 2012). SUMO conjugation is reversible and ULPs catalyse de‐conjugation. Plant ULPs form at least four subgroups that are conserved across angiosperms and function non‐redundantly (Conti et al., 2008; Novatchkova et al., 2012).
In many eukaryotes, such as budding yeast (Saccharomyces cerevisiae), fruit fly (Drosophila melanogaster) and the worm Caenorhabditis elegans, SUMO is encoded by a single‐copy gene (Flotho & Melchior, 2013). Yet, mammals and Arabidopsis express up to four paralogues. The mammalian paralogues have functionally diversified, modifying distinct but overlapping protein subsets (Citro & Chiocca, 2013). At the sequence level, the mammalian SUMO2 and SUMO3 are very similar (97% sequence identity), whereas SUMO1 only shares 47% sequence identity with SUMO2/3. Functionally, the mammalian SUMO2/3 can form SUMO chains that target their substrates for degradation, whereas SUMO1 cannot (Hay, 2013). SUMO1 and SUMO2/3 also interact with different proteins non‐covalently, as they prefer slightly different SUMO interaction motifs (SIMs) in their partners (Hecker et al., 2006; Ghisletti et al., 2007; Meulmeester et al., 2008). Interestingly, the mammalian SUMO2 is essential for embryonic development, whereas SUMO3 is dispensable (Wang et al., 2014). This functional difference between SUMO2 and SUMO3 appears to be caused by differences in their expression levels, with SUMO2 being the predominant transcript.
The genome of Arabidopsis encodes eight SUMO genes that represent five distinct types (Kurepa et al., 2003; Novatchkova et al., 2004; Colby et al., 2006). Only four of these genes are expressed (Kurepa et al., 2003; Saracco et al., 2007; Budhiraja et al., 2009). From these four genes, AtSUMO1 and AtSUMO2 are closely related, sharing 89% protein sequence identity, whereas AtSUMO3 and AtSUMO5 share only 48% and 35% identity with AtSUMO1, respectively. AtSUMO1/2 appear to represent the archetype SUMOs, as they are the closest homologues of the mammalian SUMO2/3 (with 50% protein identity). Clearly, the archetype SUMOs of yeast, mammals and plants have diverged substantially at the protein sequence level since their lineages separated in evolution.
Like their mammalian counterparts, the Arabidopsis SUMO paralogues have acquired distinct expression patterns (Van den Burg et al., 2010) and biochemical properties. For example, AtSUMO1/2 are better substrates for conjugation than is AtSUMO3 (Castano‐Miquel et al., 2011). Second, AtSUMO1/2 can form SUMO chains in vitro in the presence of only SAE1/2 and SCE1 (Colby et al., 2006; Budhiraja et al., 2009). By contrast, chain formation of AtSUMO3 can only be promoted in vitro when a truncated form of the SUMO E4 ligase PIAL2 is added (Tomanov et al., 2014). Third, the known Arabidopsis ULPs display high (iso)peptidase activity to AtSUMO1/2 conjugates, but low activity to AtSUMO3 conjugates (Chosed et al., 2006; Colby et al., 2006).
The overexpression of tagged AtSUMO1 or AtSUMO2 variants causes the activation of plant immunity, reduced rosette size and altered flowering time (Budhiraja et al., 2009; Van den Burg et al., 2010). This suggests that enhanced SUMO levels caused by gene duplication of the archetype SUMOs potentially result in a fitness cost in plants. A key question is how novel SUMO paralogues have emerged with this evolutionary penalty. Here, we report how the plant SUMO family has expanded and diversified in plants, focusing on Brassicaceae (a eudicot family) and Poaceae (a monocot family).
The genome evolution of flowering plants has been massively shaped by palaeoploidy events (Van de Peer et al., 2009). For example, one of the largest clades of angiosperms, eudicots, is characterized by an ancient whole‐genome triplication (hereafter called WGT At‐γ) that predates the split of the eudicot clades Asterids, Caryophyllales and Rosids (Tang et al., 2008; Dohm et al., 2014). Numerous gene duplicates and duplication blocks have been retained from this pan‐eudicot WGT across extant eudicots. Subsequently, two additional whole‐genome duplications (WGDs) (At‐β (88–81 million yr ago (Ma)) and At‐α (47 Ma)) occurred in the Brassicales lineage, which comprises the family Brassicaceae (Vision et al., 2000; Hohmann et al., 2015). These three palaeopolyploidy events would already have given rise to 12 gene copies in Arabidopsis for any SUMO copy present in the ancestral species that underwent At‐γ. Importantly, extensive genome synteny remains from these polyploidy events, both between and within eudicot genomes. We used this genome collinearity (i.e. correlated gene arrangements between genomic regions within and between genomes) to infer ancestry for each of the Brassicaceae SUMO genes.
An important model for gene evolution on WGDs is the dosage balance model, based on the notion that retained duplicates tend to be balanced in dosage with each other (Birchler & Veitia, 2007). The dosage balance model per se does not address mechanisms of neofunctionalization and, as such, the birth of novel UBL types, although the preservation of duplicates is an essential first step for the birth of novel UBL types (Guo et al., 2013). In agreement with this model, we reasoned that WGDs will, at first, not imbalance SUMO homeostasis, as the entire (de)conjugation machinery is duplicated. Purifying selection can then be relaxed on one duplicate, allowing it to acquire mutations. Once selection pressure is relaxed, many WGD duplicates are known to be lost. Alternatively, in unique cases, an altered function could be acquired that is beneficial. This could become fixed and then be subject to purifying selection. Our data indicate that this evolutionary model for polyploidy best explains the expansion of the Arabidopsis SUMOs, including neofunctionalization, subfunctionalization and the birth and death of novel paralogues.
Materials and Methods
Plant SUMO and SUMO‐like (SUL) sequences
Coding sequences of SUMO genes were retrieved from whole‐genome and transcriptome assemblies using BLAST searches with the Arabidopsis SUMO genes as input sequence. We used Brad (http://brassicadb.org/), Phytozome 10.1 (DOE‐JG, www.phytozome.net) and CoGe (https://genomevolution.org/) as sources (Supporting Information Table S1). The different Brassicaceae SUMO homologues were assigned to five groups on the basis of the types identified previously in A. thaliana (Kurepa et al., 2003), and multiple sequence alignments (MSAs) were made for these orthogroups. The accession numbers of the Brassicaceae SUMO genes are listed in Table S2. Support for the expression of different Brassicaceae gene models came from publically deposited transcriptomic data. For several Brassicaceae species, we have not included gene IDs in Table S2, as their assemblies lacked gene models. SUMO sequences from Cleome gyandra, Boechera stricta, Raphanus species (http://bioinfo.bti.cornell.edu/cgi-bin/radish/index.cgi), Brassica napus, Chorispora bungeana and Schrenkiella parvula (syn. Eutrema parvulum) were also retrieved from the NCBI whole‐genome and transcriptome shot gun assemblies. Brassica oleracea transcripts were retrieved from an expressed sequence tag (EST) collection (http://brassica.jcvi.org/cgi-bin/brassica/gbrowse.cgi). Amborella trichopoda sequences were retrieved from its genome assembly (www.amborella.org/) (Amborella Genome Project, 2013). MSAs were made using Muscle (http://www.ebi.ac.uk/Tools/msa/muscle/). Gene models and alignments were manually corrected using BioEdit (http://www.mbio.ncsu.edu/bioedit/page2.html). Sequences with poor coverage or quality were excluded from further analysis. The SUMO sequence logos were generated with IceLogo (http://iomics.ugent.be/icelogoserver/) (Colaert et al., 2009). Thereto, we aligned 153 archetype SUMO sequences, including SUMO genes from angiosperms, gymnosperms and mosses (Selaginella moellendorffii, Sphagnum fallax, Physcomitrella patens and Marchantia polymorpha).
Gene tree construction
Gene trees were constructed using a maximum likelihood (ML) approach in RaxML (v.7.4.12) with default settings and a GTR + gamma nucleotide model (Stamatakis et al., 2008). ML analyses were run on Cipres (http://www.phylo.org/) and the best scoring tree is shown with bootstrap support values at the nodes (Miller et al., 2010a). Tree construction for the archetype SUMO genes from eudicots was based on 109 aligned sequences with a length of 384 nucleotides (259 differential patterns); the species used are indicated in Table S1. The SUMO5 tree was reconstructed using 41 aligned sequences (nucleotide length of 241, with 209 differential patterns): Tarenaya hassleriana, Aethionema arabicum, Arabidopsis thaliana, A. lyrata, A. halleri, A. arenosa, Boechera stricta, Capsella rubella, Camelina sativa, Chorispora bungeana, Brassica rapa, B. oleracea, Raphanus raphanistrum, R. sativus, Eutrema salsugineum, Arabis alpina, Leavenworthia alabamica, Neslia paniculata, Schrenkiella parvula and Sisymbrium irio. We removed, in this case, poorly aligned regions from both the N‐ and C‐termini. The Brassicaceae SUMO1/2 tree was inferred using 49 aligned sequences (351 nucleotides and 267 differential patterns). The pruned Brassicaceae phylogeny tree was based on published data (Couvreur et al., 2010; Franzke et al., 2011; Haudry et al., 2013; Moghe et al., 2014).
Clustering analysis of the syntenic gene pairs
For the synteny‐based approach, we retrieved syntenic gene pairs (between and within genomes) using genome collinearity. Gene pairs were retrieved from the Plant Genome Duplication Database (PGDD) (http://chibba.agtec.uga.edu/duplication/) (Lee et al., 2013). The accession numbers of the dicot SUMO/SUL genes obtained are listed in Table S3. These gene pairs were represented in a network in Cytoscape (Cline et al., 2007) using the Files ‘Organic’ network lay‐out. The network representation was manually optimized to depict the three major gene SUMO/SUL clusters and to highlight their interaction with the Brassicaceae SUMO paralogues. Support for the network organization is based on the number of syntenic gene pairs between twinned genomic blocks and the scores provided for these blocks by PGDD. Edge thickness represents log(number of anchors), but a similar representation was obtained with log(score). As Tarenaya hassleriana (syn. Cleome spinosa) and Aethionema arabicum are not represented in PGDD, we performed, for these species, separate GeVo analyses in CoGe (https://genomevolution.org/coge/GEvo.pl) to obtain syntenic paralogous relationships between the SUMO genes from T. hassleriana, A. arabicum, Arabidopsis and eucalyptus (Eucalyptus grandis).
SUMO gene evolution in the Arabidopsis population
Sequence conservation of the Arabidopsis SUMO genes was assessed using the data from the 444 Arabidopsis accessions sequenced (http://signal.salk.edu/atg1001/3.0/gebrowser.php). We determined the percentage of accessions that contained an amino acid other than the prevalent residue for each position for the eight Arabidopsis SUMO paralogues. Subsequently, we aligned the SUMO paralogues in a protein MSA. We then generated a heat map of the MSA depicting the percentage of accessions (%) containing a different residue at a particular position in the MSA for each position in the alignment. The heat map was generated in R (http://www.r-project.org) using Heatmap.2 (gplots package) with the grey2yellow colour key.
Results
To reconstruct the evolution of the Arabidopsis SUMO paralogues, we searched for homologues of the five Arabidopsis SUMO types (AtSUMO1/2, AtSUMO3, AtSUMO4/6, AtSUMO5 and AtSUMO7/8) in plant genome assemblies. We always identified at least one close homologue of AtSUMO1/2 in each plant genome analysed, but close homologues were absent for the other SUMO types outside the Brassicaceae family. We only found one exception to this rule, that is, we found a SUMO5 orthologue (Th15853) in T. hassleriana; this species belongs to the closest sister family of Brassicaceae: Cleomaceae (Cheng et al., 2013). This implies that: (1) the Arabidopsis SUMO paralogues other than SUMO1/2 first emerged in a common ancestor of Brassicaceae/Cleomaceae; and (2) SUMO1/2 represents the archetype SUMO in plants. The protein sequence of these archetype SUMO homologues proved to be extremely conserved from mosses to angiosperms, specifically across the β‐grasp fold (Ala16–Gly93 for AtSUMO1) (Fig. 1a). C‐terminal to the diGly motif, the sequence is not conserved, whereas N‐terminal to the β‐grasp fold, a second motif was found to be conserved (Fig. 1b). This six‐residue motif probably acts as an internal SUMO acceptor site (QEE[D/E]KK*P, with * indicating the acceptor Lys); at least in vitro this Lys acts as a SUMO acceptor site (Colby et al., 2006). This acceptor motif is retained from mosses (P. patens, S. fallax and M. polymorpha) to angiosperms with a variant motif in S. moellendorffii (DVKPEKKP). Mosses like P. patens split c. 500 Ma from the lineage, leading to angiosperms (Hedges et al., 2015). Combined, this indicates that the archetype SUMO protein is extremely conserved in land plants and that SUMO chain formation is potentially as well conserved.
Figure 1.
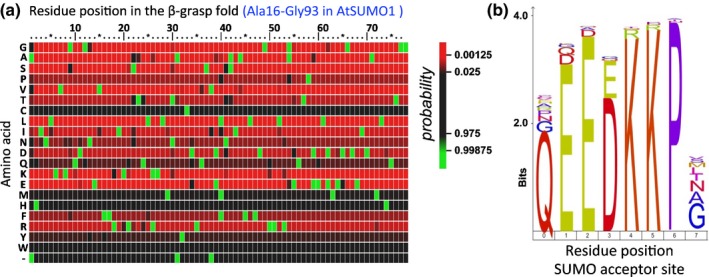
The sequences of the β‐grasp fold and the Small Ubiquitin‐Like Modifier (SUMO) acceptor motif are conserved across land plants. (a) A heat map diagram of the protein sequence alignment of archetype SUMOs from land plants, demonstrating extreme sequence conservation across the entire β‐grasp fold; of 77 positions in the β‐grasp fold, 69 positions (90%) are nearly invariant and, for the other eight positions, we observed predominantly substitutions of the presumed ancestral state for a similar residue: 25[KR], 29[TN], 39[MFL], 41[AS], 53[GA] and 38/58/61[DE]. The colour indicates the probability of a certain amino acid at that position. (b) IceLogo of the SUMO acceptor site in the N‐terminus of SUMO shows that the motif is strictly conserved in land plants. The same set of sequences is used as in (a).
Only one of two ancient archetype SUMO genes of eudicots is retained in Brassicaceae
Subsequently, we examined the moment of birth of the Arabidopsis SUMO1 and SUMO2 genes. Interestingly, we only found one SUMO gene (ID: AmTr_v1.0_scaffold00228: 122 523–131 678 bp) in the genome of the basal angiosperm Amborella trichopoda (Amborella Genome Project, 2013). Amborella trichopoda forms an outgroup to most other extant angiosperms (with an estimated split at c. 147 Ma), including the monocots and dicots. By contrast, most monocot and dicot genomes analysed contained extra SUMO gene copies (Table S1). This indicates that a common ancestor of the angiosperms potentially carried a single SUMO gene and that, during monocot and eudicot radiation, this ancestral gene was duplicated.
Based on this notion, we constructed an ML gene tree for a set of Brassicaceae SUMO1/2 genes and a core set of archetype SUMO genes from eudicot genomes other than Brassicaceae; this set included sequences from both Rosids and Asterids (Fig. 2). As outgroup for this tree, we used SUMO homologues of monocots (grasses and banana (Musa acuminata)). The gene tree revealed the existence of two major SUMO clades in eudicots (Fig. 2a). SUMO proteins in Clade A are recognizable by a variable stretch of glycines, which starts at position + 4 from the translational start; this stretch of glycines is absent in the Clade B SUMO genes. Importantly, AtSUMO1 and AtSUMO2 both grouped with Clade B. In fact, all Brassicaceae SUMO genes grouped with Clade B, whereas the archetype SUMO genes from T. hassleriana split over both clades (Fig. 2b). This indicates that Clade A was recently lost in Brassicaceae since the split with Cleomaceae (c. 52 Ma). In agreement, we found that both clades are represented in the genome of papaya (Carica papaya). Papaya represents a basal Brassicales that separated before the At‐β WGD. Also, in the genomes of sweet orange (Citrus × sinensis) and cacao (Theobroma cacao), both SUMO clades are represented (Fig. 2c). Sweet orange and cacao belong to sister orders of Brassicales, namely Sapindales and Malvales (Hohmann et al., 2015; Magallon et al., 2015). Both clades are also represented in the genomes of eucalyptus and grape (Vitis vinifera); both of these species belong to basal Eurosid lineages. In fact, both SUMO clades were also present in Asterids, for example, potato (Solanum tuberosum), tomato (S. lycopersicum) and kiwi (Actinidia chinensis). Combined, this means that at least two archetype SUMO genes have coexisted for > 125 million yr in many eudicots, but that one copy was lost specifically in a common ancestor of the Brassicaceae family.
Figure 2.
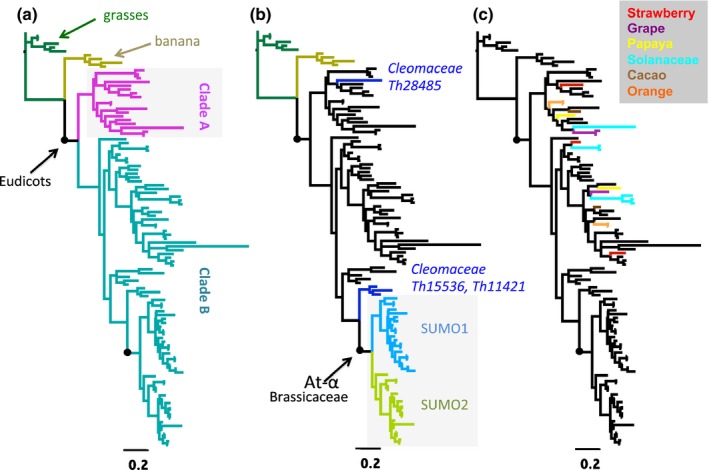
In a common ancestor of eudicots, the archetype Small Ubiquitin‐Like Modifier (SUMO) gene was duplicated with both copies being broadly retained, except for Brassicaceae. (a) Gene tree diagram of eudicot SUMO genes demonstrates that they split into two distinct clades (clades A and B). SUMO homologues from grasses (Poaceae) and banana were used as outgroup. (b) Same tree as in (a). Brassicaceae SUMO1 and SUMO2 cluster uniquely with clade B, whereas archetype SUMOs from Tarenaya hassleriana split over both clades. Tarenaya hassleriana belongs to the nearest sister family of Brassicaceae: Cleomaceae. The indicated gene IDs come from the T. hassleriana genome assembly. The Brassicaceae‐specific At‐α polyploidy event is indicated on the branch. (c) Same tree as in (a). Both clades first emerged in a common ancestor of eudicots, as they are both represented in Asterids (Solanaceae species tomato and potato (cyan)) and Eurosids (strawberry (red), grape vine (purple), papaya (yellow), cacao (brown) and sweet orange (orange)).
The Arabidopsis SUMO1 and SUMO2 genes are recent At‐α duplicates
We found that orthologues of AtSUMO1 and AtSUMO2 are present in each Brassicaceae genome analysed, including Aethionema arabicum. This species represents a basal Brassicaceae lineage that split before radiation of the Brassicaceae crown group (c. 32 Ma) (Beilstein et al., 2010; Kagale et al., 2014; Hohmann et al., 2015). We noted little protein sequence variation between AtSUMO1/2 and their Brassicaceae orthologues. The AtSUMO1/2 genes are syntenic paralogues, that is, they are located in a duplicated genomic block consisting of 90 syntenic genes (homologous gene pairs that are arranged in a related order on both genomic blocks). This duplication block was present in all Brassicaceae analysed and carries an At‐α signature, that is, the mean synonymous substitution value per synonymous site (Ks) of this duplication block (mean Ks ± SD = 0.91 ± 0.32; 90 gene anchors) corresponds to the mean Ks of the At‐α duplication blocks combined (Ks = 0.77) and not to the mean Ks of the At‐β blocks (Ks = 2.05) (Fig. S1) (Kagale et al., 2014). At‐α is absent in T. hassleriana (family Cleomaceae). Instead, T. hassleriana has experienced its own WGT (Th‐α) (Cheng et al., 2013). In agreement with this, we see that the Clade B SUMO genes of T. hassleriana form a separate branch (Th15536, Th11421) in the ML tree, which is positioned sister to the Brassicaceae SUMO1 and SUMO2 branches (Fig. 2b; grey box). From this analysis, we conclude that this AtSUMO1/2 duplication emerged as a result of At‐α and that both genes have been retained across Brassicaceae ever since.
SUMO5 appears to have neofunctionalized in Brassicaceae
We also analysed the sequence variation of the eight Arabidopsis SUMO paralogues in 444 accessions. We found a substantial number of alleles that contained non‐synonymous mutations for the four pseudogenes (AtSUMO4, AtSUMO6, AtSUMO7 and AtSUMO8). Similarly, many coding mutations were found for AtSUMO3, affecting its entire protein coding sequence (Fig. 3a). However, AtSUMO1 and AtSUMO2, but also AtSUMO5, were practically invariant at the protein level in the Arabidopsis population. For AtSUMO2, one non‐synonymous mutation was found that was present in 36 accessions, affecting the processed C‐terminal tail (F101V). Other mutations, which affect the mature AtSUMO2 protein, were only found in unique accessions (Fig. 3b). For AtSUMO1, only two accessions carried a non‐synonymous mutation (A3S).
Figure 3.
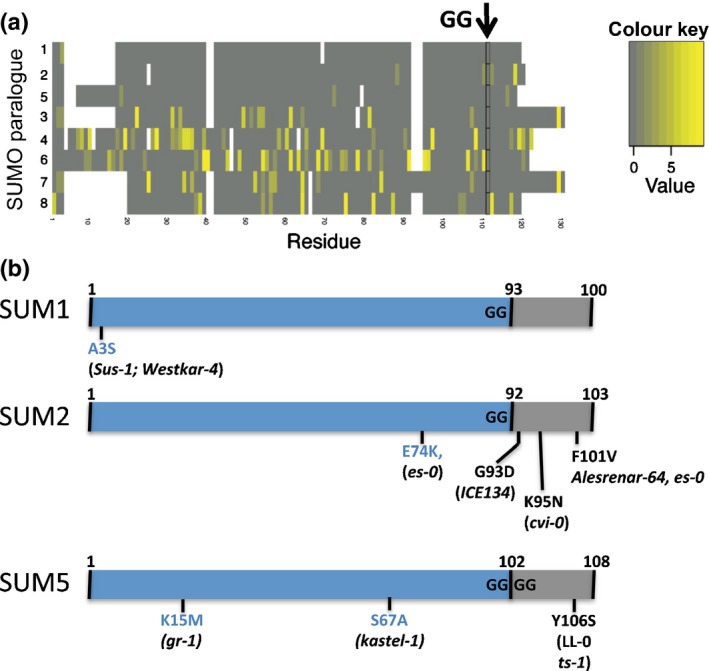
The protein sequences of AtSUMO1, AtSUMO2 and AtSUMO5 are highly conserved in the Arabidopsis population. (a) Heat map diagram of a protein sequence alignment displaying the percentage of amino acid substitutions per residue (grey to yellow) for the different Arabidopsis Small Ubiquitin‐Like Modifier (SUMO) paralogues. AtSUMO1, AtSUMO2 and AtSUMO5 have a few dominant alleles in the population with few accessions carrying an amino acid substitution, whereas, for the other paralogues, many alleles exist in the population with numerous substitutions scattered over the encoded proteins. The diglycine (diGly) motif is indicated (black arrow). The white interruptions indicate gaps in the alignment. (b) Diagram of the three conserved Arabidopsis SUMO paralogues with the substitutions found in the different accessions indicated. Blue, the mature protein; grey, the C‐terminal part removed during processing.
In the case of AtSUMO5, four accessions contained an allele that encoded an amino acid substitution compared with its sequence in the accession Col‐0. Orthologues of SUMO5 are conserved across Brassicaceae, including A. arabicum, but are more divergent than the SUMO1/2 orthologues (Fig. 4; based on branch lengths). Gene expression data (EST and whole transcriptome data) indicate that many SUMO5 orthologues are expressed (Table S2). Several Brassicaceae SUMO5 transcripts (Bra005558, Bra021812, Thhalv10015519) already encode a mature SUMO protein with three glycines exposed at the C‐terminus, indicating that processing would not be needed for these variants. Importantly, there is a close homologue of SUMO5 in T. hassleriana (Th15853), but not in the more basal Brassicales papaya. SUMO5 must therefore have evolved prior, but relatively close to, the split of Brassicaceae and Cleomaceae (Kagale et al., 2014). Since then, SUMO5 has potentially neofunctionalized, but future studies should reveal its function.
Figure 4.
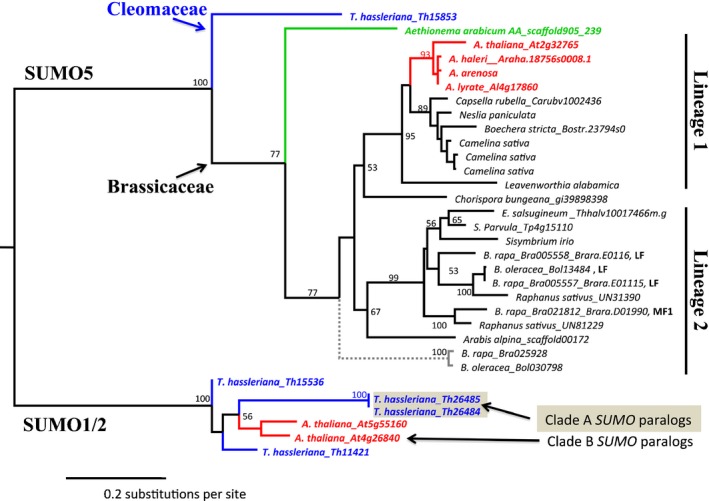
SUMO5 emerged before the split of the sister families Brassicaceae and Cleomaceae, as exemplified by Tarenaya hassleriana Th15853. The gene tree of the SUMO5 family shows that the family is less conserved than SUMO1 or SUMO2. Syntenic paralogues of SUMO5, which emerged from a Brassica‐specific whole‐genome triplication (WGT) event (Br‐α), are indicated by LF and MF1; Bra005557 + Bra005558 represent a tandem duplication. The genus Brassica also contains a putative SUMO5 pseudogene (ψ) that lacks the diglycine (diGly) motif (Bra025928, Bol03070798). As outgroup, we used SUMO1 and SUMO2 homologues of Arabidopsis and T. hassleriana; bootstrap support values are indicated for the different branches.
Identification of three ancient SUMO gene lineages in eudicots
ThSUMO5 resides in a genomic region that is syntenic with AtSUMO5, sharing 20 collinear genes (Fig. S2). This genomic region is also syntenic with a genomic region in eucalyptus, but, instead, eucalyptus contains a divergent SUL gene (Eugr.H0049, E.grandis_v1_0.046213m) at the corresponding position (Table S3). Eucalyptus belongs to the order Myrtales, a lineage that is sister to the Eurosids (Myburg et al., 2014). The split of Myrtales and Eurosids is currently estimated to have been at c. 135–110 Ma, which implies that SUMO5 evolved from a SUMO paralogue that first emerged before eudicot radiation.
To further date the birth of SUMO5, we screened for syntenic pairs of SUMO and SUL genes (using PGDD) and performed a network analysis on the gene pairs obtained using Cytoscape (Fig. 5; Table S3). This network depicts SUMO/SUL genes (nodes) that are connected by edges, which represent genome collinearity between gene pairs. The analysis revealed three major interconnected clusters of collinear genes. The two aforementioned archetype SUMO clades (Fig. 2) split perfectly over two of the three clusters, with no evidence for collinearity between them (Fig. 5). As SUMO genes from both Rosid and Asterid species are represented in both clusters, their ancestral genes must have emerged before the split of Rosids and Asterids. For example, the Rosids strawberry (Fragaria vesca), eucalyptus and grape have members in both clusters. As these three species have not undergone any additional polyploidization since At‐γ (Murat et al., 2012), these two SUMO clusters probably represent At‐γ syntenic paralogues or evolved shortly after by a gene transposition duplication event. Thereafter, homologues of both genes have been retained in many eudicots, but not in Brassicaceae (Figs 2, 5b).
Figure 5.
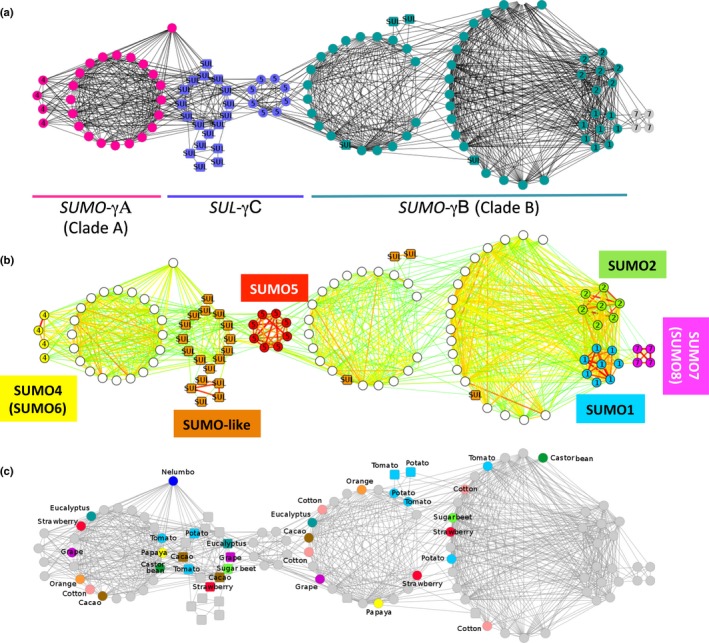
The Brassicaceae Small Ubiquitin‐Like Modifiers (SUMOs) evolved from three ancient syntenic paralogues that probably emerged from the pan‐eudicot whole‐genome triplication (WGT) (At‐γ). (a) Network representation of genome collinearity between SUMO gene pairs found within and between eudicot genomes. Two principal clusters contain the archetype SUMOs:SUMO‐γA and SUMO‐γB (purple and cyan circular nodes). These two clusters are not interconnected, but show weak synteny with a third cluster comprising SUMO5 from Brassicaceae and SUMO‐like genes from non‐Brassicaceae species (SUL‐γC; blue square nodes). The edges represent synteny between genomic regions that surround the connected genes; the numbers in the nodes refer to the Brassicaceae SUMO paralogues (1: AtSUMO1, etc.). (b) Same network as in (a), except that the Brassicaceae SUMO paralogues are indicated and the line width and colour of the edges reflect the number of syntenic genes per gene pair (green to red, low to high number of syntenic genes). Brassicaceae SUMO1 (blue) and SUMO2 (green) cluster with SUMO‐γB, whereas SUMO4 (yellow) groups with SUMO‐γA. SUMO7 (magenta) is best connected to SUMO1, whereas SUMO5 (red) is linked to both the SUMO‐γB and SUL‐γC genomic regions. (c) Same network as in (a), but the nodes are coloured per species. SUMO genes from Asterids, Rosids and Caryophyllales have representatives in each of the principal three clusters (SUMO‐γA, SUMO‐γB and SUL‐γC). The basal dicot sacred lotus (Nelumbo nucifera) is represented by a single SUMO gene, which shows synteny with both SUMO‐γA and SUL‐γC clusters, but not SUMO‐γB.
The third cluster identified comprises a set of divergent SUL sequences. We estimate the birth of this third cluster also at/or around At‐γ, because Rosids, Asterids and Caryophyllales (sugar beet (Beta vulgaris)) have members in this third cluster. The moment of birth of these three clusters is supported by the basal dicot sacred lotus (Nelumbo nucifera), which diverged from eudicots before At‐γ (Ming et al., 2013). Sacred lotus contains an archetype SUMO (NNU_022372‐RA) that shares collinearity with both the SUMO‐γA and SUL‐γC clusters, but not with the SUMO‐γB cluster. In conclusion, we found three ancient SUMO/SUL gene lineages that appear to represent At‐γ syntenic paralogues: SUMO‐γA, SUMO‐γB and SUL‐γC.
SUMO5 resides in a genomic region that acts as a hotspot for SUMO paralogue evolution in eudicots
As the SUMO5 orthologues show weak synteny to both the SUMO‐γB and SUL‐γC clusters (and no synteny with the SUMO‐γA cluster), we examined in more detail to which cluster SUMO5 belongs. Close inspection of the synteny between T. hassleriana SUMO5 (Th15853), AtSUMO5 (At2g32765) and the eucalyptus SUL gene Eucgr.H00049 (SUL‐γC) indicates that SUMO5 most probably emerged from SUL‐γC and not SUMO‐γB (Fig. S2). In agreement, the genomic block that surrounds A. lyrata SUMO5 is better connected with the SUL‐γC genes from the basal eudicots eucalyptus and grape vine than with the SUMO‐γB genes from these same species (Table S4). Another argument that SUMO5 emerged from SUL‐γC is that the genes in this cluster appear to diverge, that is, the SUL‐γC sequences do not form a gene tree that is consistent with their species tree. By contrast, the two other clusters contain primarily close homologues of the archetype SUMO and their sequences diverge little. We therefore propose that SUMO5 most probably emerged from a SUL‐γC predecessor.
Origin of the Brassicaceae SUMO4 and SUMO7 orthogroups
The four Arabidopsis SUMO pseudogenes are arranged as two tandem duplications (TDs), that is, AtSUMO4::AtSUMO6 (At5g48700, At5g48710) and AtSUMO7::AtSUMO8 (At5g5‐5855, At5g55856) (Kurepa et al., 2003). However, in other Brassicaceae – including the genus Arabidopsis (A. halleri, A. arenosa and A. lyrata), they are present as singletons at syntenic scaffolds. Therefore, both TDs probably occurred during A. thaliana speciation. Interestingly, AtSUMO4 shares collinearity with SUMO‐γA genes, including eucalyptus Eucgr.H00789 and the T. hassleriana TD gene pair Th26484 and Th26485 (Figs 5, S3; Table S3). For example, we found 25 collinear genes between Arabidopsis and T. hassleriana. This indicates that AtSUMO4 and Th26484::Th26485 are syntenic orthologues. SUMO4 is also present in A. arabicum and other Brassicaceae (Table S2). This means that AtSUMO4 must have emerged in a common ancestor of Brassicaceae after the split of Cleomaceae, and that it probably evolved from a SUMO‐γA descendant.
Related to this, we noted that Brassicaceae SUMO7 shows collinearity with both Brassicaceae SUMO1/2, but not with SUMO4 or SUMO5 (Fig. 5b). The average Ks between the SUMO7 and SUMO1/2 genomic regions is c. 1.0–1.1. This is more than expected for At‐α (Ks = 0.77), but less than expected for At‐β (Ks = 2.05) (Kagale et al., 2014). This means that SUMO7 probably emerged from a segmental duplication of SUMO1 or SUMO2. The birth of SUMO7 appears to coincide with At‐α, as A. arabicum contains a putative orthologue (Table S2), but T. hassleriana does not. SUMO7 is also present in the Brassicaceae lineage II (including Brassica) (Table S2). In two Brassica species, SUMO7 is present as a misannotated singleton. In B. rapa, a homologous sequence is present in the intergenic region between Bra00287070 and Bra00287071, whereas, in B. oleracea, the corresponding gene is misannotated (Bol006236). Certain SUMO4 and SUMO7 orthologues have retained their diGly motif, whilst transcripts were also reported for SUMO4 in B. oleracea and C. bungeana, whereas, for SUMO7, a transcript was reported for C. rubella. This could mean that certain SUMO4 and SUMO7 orthologues still encode functional proteins.
SUMO3 emerged from a SUMO2 TD after divergence of A. arabicum, but before radiation of the Brassicaceae crown group
Similar to the two aforementioned Arabidopsis SUMO pseudogenes, AtSUMO2 and AtSUMO3 represent a TD (Fig. 6). This TD is present in many, but not all, Brassicaceae genomes. For example, this duplication is absent in the basal Brassicaceae A. arabicum. In E. salsugineum, two EsSUMO2 copies are present in tandem at this locus, suggesting a recent gene conversion of SUMO3. This is supported by the increased branch length of one of the two EsSUMO2 genes (*; Thhalv10015081) (Fig. S4). In the Brassica species B. rapa and B. oleracea, SUMO3 appears to be deleted from all three subgenomes (Brassica emerged from a recent ancestral hexaploid c. 20–24 Ma (Br‐α)), that is, BLAST searches did not reveal any homology to AtSUMO3. Nonetheless, SUMO3 transcripts were reported for B. oleracea (asmbl_13151; http://brassica.jcvi.org/cgi-bin/brassica/index.cgi) and B. napus (NCBI ES966440.1). The latter species is an allotetraploid of B. rapa and B. oleracea. Possibly, a single SUMO3 copy has been retained in some, but not all, Brassica cultivars. In support of this, a SUMO3 copy is retained in the genus Raphanus, which shares the Br‐α WGT and only recently diverged from the genus Brassica (5–16 Ma) (Moghe et al., 2014; Hohmann et al., 2015).
Figure 6.
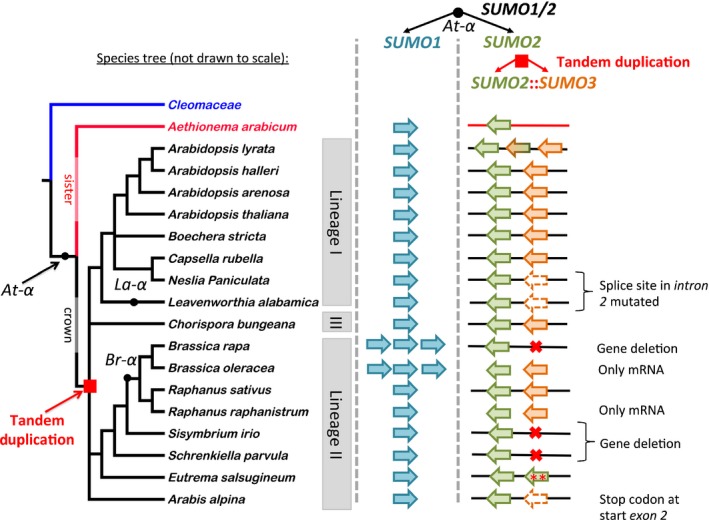
SUMO3 emerged before radiation of the Brassicaceae crown species. SUMO1 (light blue arrow) and SUMO2 (green arrow) are At‐α syntenic paralogues (black dot) based on collinearity between their genomic regions. Subsequently, a tandem duplication (TD) (red square) occurred at the SUMO2 locus (green and orange arrows) before radiation of the Brassicaceae crown group into three lineages. The phylogenetic tree represents a pruned Brassicaceae family tree with Cleomaceae (Tarenaya hassleriana) as outgroup (blue) and Aethionema arabicum (red) at the base of the Brassicaceae family tree. Various Brassicaceae lack a functional SUMO3 gene as a result of gene deletion, conversion or mutations that affect the reading frame. The Arabidopsis lyrata genome contains, in addition, a hybrid SUMO2‐3 gene.
Importantly, the ML gene tree of SUMO1, SUMO2 and SUMO3 combined indicates that SUMO2 from A. arabicum forms a branch that is basal to the SUMO2 and SUMO3 clades in the gene tree. The most parsimonious explanation is that SUMO3 emerged from a TD of SUMO2 after the split of A. arabicum (c. 32 Ma), but before radiation of the Brassicaceae crown group (Hohmann et al., 2015). On duplication, one duplicate appears to have rapidly diversified, yielding SUMO3, whereas the other duplicate remained nearly unchanged (SUMO2). The crown group is subdivided into three lineages. The split between lineage I–III and II is currently estimated at c. 23 Ma (Hohmann et al., 2015). SUMO3 orthologues are widely found and expressed in lineage I. However, in lineage II, SUMO3 is often pseudogenized (via early stop codons and mutation of intron‐splice sites), deleted or subject to gene conversion. As the two genes have co‐evolved, it is evident that SUMO2 is under purifying selection, whereas SUMO3 appears to be non‐essential in many Brassicaceae.
Birth of grass‐specific SUMO paralogues
In the genomes of grasses (Poaceae), we identified three distinct SUMO/SUL loci. Two loci are genetically linked on chromosome 2 of Brachypodium distachyon. They represent an archetype SUMO (Bradi2g58830) and, 2.7 Mb upstream, an uncharacterized grass‐specific SUMO paralogue (Bradi2g55140), hereafter called Grass SUMO‐Like 1 (GSUL1). Functional data are lacking for this GSUL1, but GSUL1 from sorghum encodes a conjugation‐deficient variant, indicating that it cannot act as PTM. In rice and maize, an orthologue of GSUL1 is missing, whereas the archetype SUMO is represented by a TD. Interestingly, this grass locus with archetype SUMO genes is related to the eudicot SUMO‐γA cluster, as the SUMO genes from banana (M. acuminata GSMUA_Achr8G00860::70 TD) and oil palm (Elaeis guineensis p5_sc00157.V1.gene38) show synteny with both the eudicot SUMO‐γA cluster and this grass locus (based on PGDD). Representatives of this grass locus are Bradi2g58830, maize (Zea mays) GRMZM2G053898, sorghum (Sorghum bicolor) Sobic.003g402600 and rice (Oryza sativa) Os01g68940. On the other hand, we found no synteny with the eudicot SUMO‐γB cluster, which supports our notion that SUMO‐γB only first appeared after the At‐γ WGT.
The third locus represents a hypervariable multi‐gene locus that contains a second grass‐specific SUL gene, hereafter called GSUL2. The locus is composed of a variable number of GSUL2 genes in different grass genomes, suggesting active gene duplications and rearrangements (Fig. 7). It is not only composed of genes with a single UBL domain, but also harbours concatemers of UBL domains. One such concatemer has been characterized previously: the maize gene DiSUMO‐like (DSUL, GRMZM2G006324; Srilunchang et al., 2010). Some of these GSUL2 genes encode conjugation‐deficient SUL proteins lacking a diGly motif, for example, Sobic.002g350100 from sorghum. Likewise, in several cases, the concatemers have lost their internal and/or C‐terminal diGly motifs, meaning that they cannot be proteolytically cleaved in conjugation‐competent single or multimeric GSUL2 units. A gene tree based on the individual UBL domains of the GSUL2 homologues, DSUL and other concatemers exposed that DSUL represents a gene fusion of two progenitor GSUL2 genes that group with two different clades in the gene tree (Figs 7a, S5). Moreover, the UBL domains from the rice concatemers also branch over two clades, but these two clades with rice UBLs do not overlap with the two DSUL clades (Fig. 7). This implies that the maize DSUL and two rice concatemers, Os07g38700, Os07g38710, emerged from independent gene fusion events. In conclusion, grasses contain an additional SUL gene cluster that actively evolves via TDs in combination with gene fusions.
Figure 7.
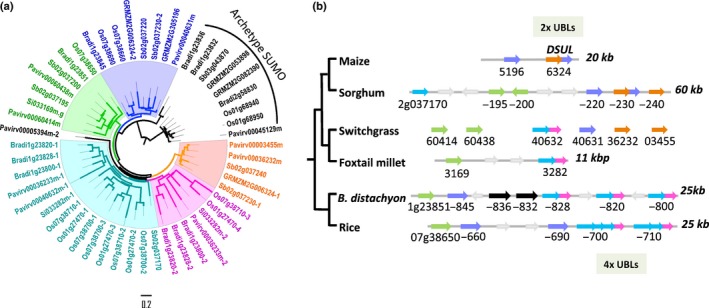
The locus in grasses that is homologous to the maize DiSUMO‐like (DSUL) locus acts as a ‘hotspot’ for SUMO‐like gene evolution, including the formation of concatemers by head‐to‐tail fusions of ubiquitin‐like modifier (UBL) domains. (a) A gene tree based on the single UBL domains from GSUL2 isoforms and UBL concatemers found at the DSUL locus in grasses. The UBL domains form five distinct clades. The UBL domains of DSUL group into different clades than the UBL domains of the rice concatemers. This implies that the gene fusions have independently occurred in ancestors of maize and rice. As outgroup, we used the archetype SUMO gene from grasses; bootstrap support values are shown in Supporting Information Fig. S5. (b) Schematic representation of the GSUL2/DSUL loci in grasses, indicating the different gene fusions and TD events found at this locus based on the maximum likelihood (ML) tree. The colours of the UBL domains (arrows) reflect the different clades seen in the ML tree. For orientation, the different gene identifiers are indicated.
Discussion
We examined the evolution and diversification of the SUMO family across angiosperms, and in greater detail in Brassicaceae and Poaceae, to understand the dynamics and evolution of novel UBLs. Expansion and divergence of the SUMO family is impacted by WGDs and TDs. The SUMO landscape is extensively shaped by the pan‐eudicot At‐γ WGT. From this WGT, three SUMO loci (SUMO‐γA, SUMO‐γB and SUL‐γC) are preserved across eudicots, of which two loci encode archetype SUMOs (SUMO‐γA, SUMO‐γB; Fig. 8). These SUMOs have remained nearly identical, suggesting that the ancestral palaeo‐eudicot SUMO genes subfunctionalized in their expression pattern or gene dosage. Importantly, the genes that belong to these two syntenic clusters split perfectly over two distinct branches in the gene tree without cases of gene conversion (Fig. 2; Table S3). We therefore rule out that they co‐evolved by concerted evolution, as reported for ubiquitin (Nei et al., 2000). From the same period, a third locus emerged that represents a diversifying orthogroup, represented by SUMO5 in Brassicaceae (Fig. 4; Table S3). Genome collinearity indicated that a SUMO5 ancestor (SUL‐γC) first emerged close to the At‐γ event (Figs 5, 8) – a moment in which the entire SUMO machinery was triplicated. Homologues derived from this ancestral SUL‐γC locus are now highly divergent between eudicot families. Hence, this genomic region acts as a hotspot for SUMO paralogue evolution. Similarly, grasses also contain a locus that acts as a hotspot for SUL evolution (Fig. 7); this locus contains both single UBL‐domain SUL genes and genes encoding concatemers of UBL domains.
Figure 8.
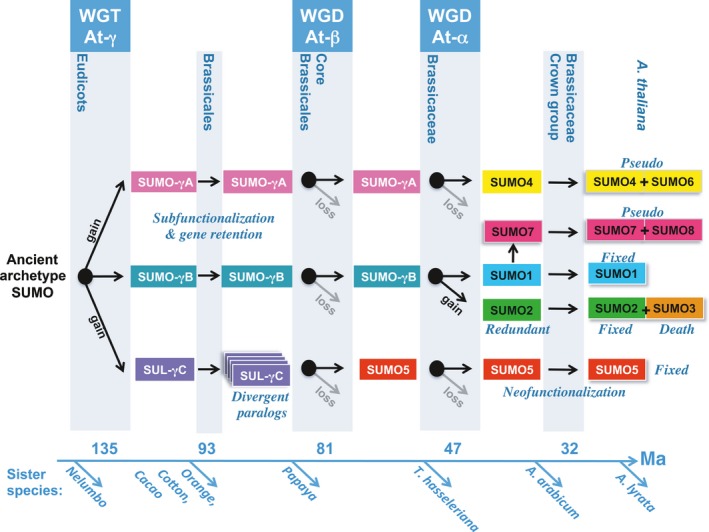
Model for the expansion and diversification of the Brassicaceae Small Ubiquitin‐Like Modifier (SUMO) gene family. The ancestral archetype SUMO gene was triplicated by the eudicot‐specific hexaploidy event At‐γ (gain). Two duplicates were broadly retained as archetype SUMOs (SUMO‐γA and SUMO‐γB) in eudicots, possibly as a result of subfunctionalization, whereas the third copy diversified shortly after (SUL‐γC) and became fixed as SUMO5 before the split of the Cleomaceae (Tarenaya hassleriana) and Brassicaceae families. Subsequently, the At‐α whole‐genome duplication (WGD) caused duplication of SUMO‐γB yielding Brassicaceae SUMO1/2. A subsequent tandem duplication of SUMO2 in a common ancestor of the Brassicaceae crown group provided SUMO3 (gain). SUMO3 is frequently lost by gene deletion, conversion or other mutations, whereas SUMO2 is 100% retained. Around this time, the SUMO‐γA homologue was pseudogenized in a recent Brassicaceae ancestor giving SUMO4. Likewise, a segmental duplication of SUMO1 or SUMO2 appears to have yielded the SUMO7 pseudogene (gain) in a recent Brassicaceae ancestor. The positions of the three most recent Arabidopsis palaeo‐polyploidy events are indicated along the top of the x‐axis (bottom). Informative sister species for this model are indicated at the bottom along the x‐axis. Ma, million years ago.
We found that the AtSUMO5 sequence is nearly invariant in Arabidopsis, which suggests that it has neofunctionalized. In agreement, the overexpression of mature AtSUMO5 resulted in its conjugation to unknown plant proteins (Budhiraja et al., 2009), indicating that it can act as PTM. Brassicaceae SUMO5 homologues have retained their diGly motif for > 52 million yr, whereas homologues of the ‘younger’ SUMO4 have frequently lost their diGly motif. Certain SUL‐γC homologues have also retained their diGly motif, suggesting that they could act as PTMs (Table S3). Biochemically, SUMO5 appears to have diverged from the canonical conjugation pathway. For example, Arabidopsis SAE1/2 and SCE2 can attach AtSUMO5 to substrates in vitro, albeit at a reduced rate compared with AtSUMO1/2 (Castano‐Miquel et al., 2011). AtSUMO5 is also a poor substrate for the known Arabidopsis ULPs (Chosed et al., 2006; Colby et al., 2006). As the birth of the SUMO5/SUL‐γC gene lineage was close to At‐γ, additional gene copies of the SUMO machinery were probably present in this ancestral species. In line with this, additional SCE1 gene copies exist in extant eudicot genomes, but not Arabidopsis (Novatchkova et al., 2012). By contrast, outside the Plant kingdom, SCE1 is mostly present as a single gene (Knobbe et al., 2015). It will be interesting to examine whether these additional SCE1 copies have co‐evolved with certain SUL genes and have composed novel conjugation pathways.
Remarkably, history has repeated itself in the case of Brassicaceae SUMO1/2, that is, they exemplify At‐α duplicates that have descended from one of the two archetype eudicot SUMO genes. SUMO1/2 appear to be strictly conserved in Brassicaceae, which implies that they act non‐redundantly and have subfunctionalization in their expression pattern. We have shown previously that this gene pair exhibits tissue‐specific gene expression in Arabidopsis (Van den Burg et al., 2010). This agrees with the notion that the loss of cis‐regulatory elements allows gene retention as a result of subfunctionalization (Haberer et al., 2004), a situation that is reminiscent of the mammalian SUMO2/3 (Wang et al., 2014). However, reverse genetics have indicated that, at least in Arabidopsis, AtSUMO1/2 act redundantly, as the knockout of either gene does not cause growth defects, whereas the double mutant is embryo lethal (Saracco et al., 2007). Overexpression of either SUMO gene triggers defence activation, whereas expression of dominant‐negative variants activates, even more strongly, plant defence (Van den Burg et al., 2010). Combined, SUMO1/2 appear to represent an example of a gene pair whose expression is dosage balance sensitive (Birchler & Veitia, 2007; De Smet et al., 2013). The cause of the dominant‐negative effect is unclear, but, in particular, ULP activity can be inhibited by SUMO overexpression (Mukhopadhyay & Dasso, 2007). By contrast, increased E2 activity via the overexpression or additional SCE1 gene copies appears not to be detrimental to plants (Novatchkova et al., 2012; Tomanov et al., 2012).
SUMO3 emerged from a TD of SUMO2 shortly after At‐α, but before radiation of the Brassicaceae crown group. This TD truncated the SUMO2 promoter (c. 381 bp in Arabidopsis, TAIR), which might be causal to SUMO2 subfunctionalization. This extra SUMO copy rapidly diverged over a short period, yielding SUMO3. Genetic studies have indicated that AtSUMO3 is not essential as the knockout is viable (Van den Burg et al., 2010). Moreover, SUMO3 is frequently deleted, converted back to SUMO2 or pseudogenized in other Brassicaceae (Fig. 6). Yet, SUMO3 appears to have neofunctionalized in Arabidopsis, as the gene product has been reported to specifically interact with the salicylic acid receptor NPR1 (Saleh et al., 2015) and its expression is transiently induced by this hormone (Van den Burg et al., 2010).
Sequence fingerprints were found for both pseudogenes SUMO4 and SUMO7 in different Brassicaceae, including A. arabicum. In fact, SUMO4 evolved from a SUMO‐γA copy in a recent ancestor of Brassicaceae, whereas SUMO7 potentially emerged from a segmental duplication involving SUMO1 in that period (Fig. 8). During this time, the Brassicaceae lineage underwent the At‐α WGD, which might have increased the SUMO gene copy number and its protein levels. As a consequence, WGD might have incited neutral selection pressure on SUMO4 and SUMO7, resulting in sequence divergence followed by their pseudogenization. Although the SUMO‐γA duplicates were lost/pseudogenized, we noted that the At‐α gene pair that emerged from the SUMO‐γB gene subfunctionalized, resulting in AtSUMO1/2. Both of these observations agree with the notion that housekeeping genes are frequently seen to revert to the singleton state, or subfunctionalize in terms of expression on WGDs (De Smet et al., 2013).
Interestingly, grasses also contain a diversifying multigene locus that encompasses a tandem array of SUMO paralogues (Srilunchang et al., 2010). Future studies should help to resolve how this DSUL/GSUL2 locus emerged. This multigene locus is subject to active TDs and gene rearrangements, resulting in functional head‐to‐tail gene fusions of SUL domains. As the number of TDs and UBL repeats in the concatemers vary between closely related species and individual UBL repeats of maize DSUL and rice concatemers group with different clades in the ML tree, it is highly likely that these genomic rearrangements and gene fusions have occurred very recently. This locus exemplifies how di‐ubiquitin‐like proteins ISG15, FAT10 and RUB1 might have evolved in various eukaryotes (Mergner & Schwechheimer, 2014; Basler et al., 2015; Radoshevich et al., 2015). Related to this, it has been reported that SCE1 is duplicated in grasses (Novatchkova et al., 2012) and that two distinct phylogenetic subclades are retained, suggesting that GSUL1, GSUL2 and/or DSULs could potentially have co‐evolved with this divergent SCE1 orthogroup in monocots.
We have found that, in plants, WGDs followed by TDs are important drivers for SUMO paralogue evolution. For example, the pan‐eudicot palaeohexaploidy event has yielded a widespread locus that acts as ‘hotspot’ for SUMO paralogue evolution in eudicots, whereas, in Brassicaceae, the paralogue SUMO3 only emerged after a WGD followed by a TD of one duplicate. Despite these cases of paralogue evolution, we have found that the SUMO gene copy number appears to have reverted to a singleton state in plants, and the retained archetype SUMOs have subfunctionalized in terms of their expression pattern and not in terms of their sequence.
Author contributions
H.A.vdB. and M.E.S. designed the research. H.A.vdB. and V.H. carried out data analysis and interpretation. H.A.vdB., V.H. and G.V. performed the research. H.A.vdB., V.H. and M.E.S. wrote the manuscript.
Supporting information
Please note: Wiley Blackwell are not responsible for the content or functionality of any supporting information supplied by the authors. Any queries (other than missing material) should be directed to the New Phytologist Central Office.
Fig. S1 Frequency distribution of the mean synonymous substitution value per synonymous site (Ks) for the retained gene duplicates in the AtSUMO1 and AtSUMO2 duplication block.
Fig. S2 Brassicaceae SUMO5 evolved from an ancient Small Ubiquitin‐Like Modifier (SUMO)‐like paralog found in eudicots.
Fig. S3 Brassicaceae SUMO4 originates from an archetype Small Ubiquitin‐Like Modifier (SUMO) that diversified after the split of Brassicaceae and Cleomaceae.
Fig. S4 Maximum likelihood (ML) tree of Brassicaceae SUMO1, SUMO2 and SUMO3 genes, indicating that Aethionema arabicum SUMO2 groups sister to the SUMO2 and SUMO3 clades.
Fig. S5 Gene tree of the individual ubiquitin‐like modifier (UBL) domains of the DiSUMO‐like (DSUL) locus in grasses with gene identifiers and bootstrap support values indicated.
Table S1 List of the different plant genomes used in this study
Table S2 Annotation of the Brassicaceae Small Ubiquitin‐Like Modifier (SUMO) paralogues, including expression details.
Table S3 Gene IDs of the dicot Small Ubiquitin‐Like Modifier/SUMO‐like (SUMO/SUL) genes studied here in the three different genomic regions: SUMO‐γA (AtSUMO4), SUMO‐γB (AtSUMO1/2) and SUL‐γC (AtSUMO5)
Table S4 Summary of the synteny between Arabidopsis lyrata SUMO5 (ID: 16062200; Al4g17860) and the Small Ubiquitin‐Like Modifier (SUMO) and SUMO‐like (SUL) genes of the basal eudicots grape vine (Vitis vinifera) and eucalyptus (Eucalyptus grandis)
Acknowledgements
The Netherlands Scientific Organisation supported this work (ALW‐VIDI 864.10.004 to H.A.vdB.). We are grateful to George Coupland and Geo Velikkakam (MPI, Cologne), and Luca Comai (UC Davis Genome Center), who kindly provided access to sequence assemblies for Arabis alpina and Arabidopsis arenosa, respectively. Christa Testerink and Paul Fransz (University of Amsterdam) kindly provided Arabidopsis accessions. Freek Bakker (Wageningen University), Frank Takken and Like Fokkens (University of Amsterdam) are thanked for providing critical reviews and helpful comments.
References
- Amborella Genome Project . 2013. The Amborella genome and the evolution of flowering plants. Science 342: 1241089. [DOI] [PubMed] [Google Scholar]
- Basler M, Buerger S, Groettrup M. 2015. The ubiquitin‐like modifier FAT10 in antigen processing and antimicrobial defense. Molecular Immunology 68: 129–132. [DOI] [PubMed] [Google Scholar]
- Beilstein MA, Nagalingum NS, Clements MD, Manchester SR, Mathews S. 2010. Dated molecular phylogenies indicate a Miocene origin for Arabidopsis thaliana . Proceedings of the National Academy of Sciences, USA 107: 18724–18728. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Birchler JA, Veitia RA. 2007. The gene balance hypothesis: from classical genetics to modern genomics. Plant Cell 19: 395–402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Budhiraja R, Hermkes R, Muller S, Schmidt J, Colby T, Panigrahi K, Coupland G, Bachmair A. 2009. Substrates related to chromatin and to RNA‐dependent processes are modified by Arabidopsis SUMO isoforms that differ in a conserved residue with influence on desumoylation. Plant Physiology 149: 1529–1540. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Burroughs AM, Iyer LM, Aravind L. 2012. Structure and evolution of ubiquitin and ubiquitin‐related domains. Methods in Molecular Biology 832: 15–63. [DOI] [PubMed] [Google Scholar]
- Callis J. 2014. The ubiquitination machinery of the ubiquitin system. Arabidopsis Book 12: e0174. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Castano‐Miquel L, Segui J, Lois LM. 2011. Distinctive properties of Arabidopsis SUMO paralogues support the in vivo predominant role of AtSUMO1/2 isoforms. Biochemical Journal 436: 581–590. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Castano‐Miquel L, Segui J, Manrique S, Teixeira I, Carretero‐Paulet L, Atencio F, Lois LM. 2013. Diversification of SUMO‐activating enzyme in Arabidopsis: implications in SUMO conjugation. Molecular Plant 6: 1646–1660. [DOI] [PubMed] [Google Scholar]
- Cheng S, van den Bergh E, Zeng P, Zhong X, Xu J, Liu X, Hofberger J, de Bruijn S, Bhide AS, Kuelahoglu C et al 2013. The Tarenaya hassleriana genome provides insight into reproductive trait and genome evolution of crucifers. Plant Cell 25: 2813–2830. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chosed R, Mukherjee S, Lois LM, Orth K. 2006. Evolution of a signalling system that incorporates both redundancy and diversity: Arabidopsis SUMOylation. Biochemical Journal 398: 521–529. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Citro S, Chiocca S. 2013. Sumo paralogs: redundancy and divergencies. Frontiers in Bioscience 5: 544–553. [DOI] [PubMed] [Google Scholar]
- Cline MS, Smoot M, Cerami E, Kuchinsky A, Landys N, Workman C, Christmas R, Avila‐Campilo I, Creech M, Gross B et al 2007. Integration of biological networks and gene expression data using Cytoscape. Nature Protocols 2: 2366–2382. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Colaert N, Helsens K, Martens L, Vandekerckhove J, Gevaert K. 2009. Improved visualization of protein consensus sequences by iceLogo. Nature Methods 6: 786–787. [DOI] [PubMed] [Google Scholar]
- Colby T, Matthai A, Boeckelmann A, Stuible HP. 2006. SUMO‐conjugating and SUMO‐deconjugating enzymes from Arabidopsis. Plant Physiology 142: 318–332. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Conti L, Price G, O'Donnell E, Schwessinger B, Dominy P, Sadanandom A. 2008. Small ubiquitin‐like modifier proteases OVERLY TOLERANT TO SALT1 and ‐2 regulate salt stress responses in Arabidopsis . Plant Cell 20: 2894–2908. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Couvreur TL, Franzke A, Al‐Shehbaz IA, Bakker FT, Koch MA, Mummenhoff K. 2010. Molecular phylogenetics, temporal diversification, and principles of evolution in the mustard family (Brassicaceae). Molecular Biology and Evolution 27: 55–71. [DOI] [PubMed] [Google Scholar]
- De Smet R, Adams KL, Vandepoele K, Van Montagu MC, Maere S, Van de Peer Y. 2013. Convergent gene loss following gene and genome duplications creates single‐copy families in flowering plants. Proceedings of the National Academy of Sciences, USA 110: 2898–2903. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dohm JC, Minoche AE, Holtgrawe D, Capella‐Gutierrez S, Zakrzewski F, Tafer H, Rupp O, Sorensen TR, Stracke R, Reinhardt R et al 2014. The genome of the recently domesticated crop plant sugar beet (Beta vulgaris). Nature 505: 546–549. [DOI] [PubMed] [Google Scholar]
- Flotho A, Melchior F. 2013. Sumoylation: a regulatory protein modification in health and disease. Annual Review of Biochemistry 82: 357–385. [DOI] [PubMed] [Google Scholar]
- Franzke A, Lysak MA, Al‐Shehbaz IA, Koch MA, Mummenhoff K. 2011. Cabbage family affairs: the evolutionary history of Brassicaceae. Trends in Plant Sciences 16: 108–116. [DOI] [PubMed] [Google Scholar]
- Ghisletti S, Huang W, Ogawa S, Pascual G, Lin ME, Willson TM, Rosenfeld MG, Glass CK. 2007. Parallel SUMOylation‐dependent pathways mediate gene‐ and signal‐specific transrepression by LXRs and PPARgamma. Molecular Cell 25: 57–70. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guo H, Lee TH, Wang X, Paterson AH. 2013. Function relaxation followed by diversifying selection after whole‐genome duplication in flowering plants. Plant Physiology 162: 769–778. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Haberer G, Hindemitt T, Meyers BC, Mayer KF. 2004. Transcriptional similarities, dissimilarities, and conservation of cis‐elements in duplicated genes of Arabidopsis. Plant Physiology 136: 3009–3022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Haudry A, Platts AE, Vello E, Hoen DR, Leclercq M, Williamson RJ, Forczek E, Joly‐Lopez Z, Steffen JG, Hazzouri KM et al 2013. An atlas of over 90,000 conserved noncoding sequences provides insight into crucifer regulatory regions. Nature Genetics 45: 891–898. [DOI] [PubMed] [Google Scholar]
- Hay RT. 2013. Decoding the SUMO signal. Biochemical Society Transactions 41: 463–473. [DOI] [PubMed] [Google Scholar]
- Hecker CM, Rabiller M, Haglund K, Bayer P, Dikic I. 2006. Specification of SUMO1‐ and SUMO2‐interacting motifs. Journal of Biological Chemistry 281: 16117–16127. [DOI] [PubMed] [Google Scholar]
- Hedges SB, Marin J, Suleski M, Paymer M, Kumar S. 2015. Tree of life reveals clock‐like speciation and diversification. Molecular Biology and Evolution 32: 835–845. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hohmann N, Wolf EM, Lysak MA, Koch MA. 2015. A time‐calibrated road map of Brassicaceae species radiation and evolutionary history. Plant Cell 27: 2770–2789. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang L, Yang S, Zhang S, Liu M, Lai J, Qi Y, Shi S, Wang J, Wang Y, Xie Q et al 2009. The Arabidopsis SUMO E3 ligase AtMMS21, a homologue of NSE2/MMS21, regulates cell proliferation in the root. Plant Journal 60: 666–678. [DOI] [PubMed] [Google Scholar]
- Ishida T, Fujiwara S, Miura K, Stacey N, Yoshimura M, Schneider K, Adachi S, Minamisawa K, Umeda M, Sugimoto K. 2009. SUMO E3 ligase HIGH PLOIDY2 regulates endocycle onset and meristem maintenance in Arabidopsis. Plant Cell 21: 2284–2297. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ishida T, Yoshimura M, Miura K, Sugimoto K. 2012. MMS21/HPY2 and SIZ1, two Arabidopsis SUMO E3 ligases, have distinct functions in development. PLoS ONE 7: e46897. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jentsch S, Psakhye I. 2013. Control of nuclear activities by substrate‐selective and protein‐group SUMOylation. Annual Review of Genetics 47: 167–186. [DOI] [PubMed] [Google Scholar]
- Kagale S, Robinson SJ, Nixon J, Xiao R, Huebert T, Condie J, Kessler D, Clarke WE, Edger PP, Links MG et al 2014. Polyploid evolution of the Brassicaceae during the Cenozoic era. Plant Cell 26: 2777–2791. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Knobbe AR, Horken KM, Plucinak TM, Balassa E, Cerutti H, Weeks DP. 2015. SUMOylation by a stress‐specific small ubiquitin‐like modifier E2 conjugase is essential for survival of Chlamydomonas reinhardtii under stress conditions. Plant Physiology 167: 753–765. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kurepa J, Walker JM, Smalle J, Gosink MM, Davis SJ, Durham TL, Sung DY, Vierstra RD. 2003. The small ubiquitin‐like modifier (SUMO) protein modification system in Arabidopsis. Journal of Biological Chemistry 278: 6862–6872. [DOI] [PubMed] [Google Scholar]
- Lee TH, Tang H, Wang X, Paterson AH. 2013. PGDD: a database of gene and genome duplication in plants. Nucleic Acids Research 41: D1152–D1158. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Magallon S, Gomez‐Acevedo S, Sanchez‐Reyes LL, Hernandez‐Hernandez T. 2015. A metacalibrated time‐tree documents the early rise of flowering plant phylogenetic diversity. New Phytologist 207: 437–453. [DOI] [PubMed] [Google Scholar]
- Mergner J, Schwechheimer C. 2014. The NEDD8 modification pathway in plants. Frontiers in Plant Science 5: 103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meulmeester E, Kunze M, Hsiao HH, Urlaub H, Melchior F. 2008. Mechanism and consequences for paralog‐specific sumoylation of ubiquitin‐specific protease 25. Molecular Cell 30: 610–619. [DOI] [PubMed] [Google Scholar]
- Miller MA, Pfeiffer W, Schwartz T. 2010a. Creating the CIPRES Science Gateway for inference of large phylogenetic trees. Proceedings of the Gateway Computing Environments Workshop (GCE) New Orleans, LA, USA. New York, NY, USA: IEEE, 1–8.
- Miller MJ, Barrett‐Wilt GA, Hua Z, Vierstra RD. 2010b. Proteomic analyses identify a diverse array of nuclear processes affected by small ubiquitin‐like modifier conjugation in Arabidopsis. Proceedings of the National Academy of Sciences, USA 107: 16512–16517. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miller MJ, Scalf M, Rytz TC, Hubler SL, Smith LM, Vierstra RD. 2013. Quantitative proteomics reveals factors regulating RNA biology as dynamic targets of stress‐induced SUMOylation in Arabidopsis. Molecular and Cellular Proteomics 12: 449–463. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ming R, VanBuren R, Liu Y, Yang M, Han Y, Li LT, Zhang Q, Kim MJ, Schatz MC, Campbell M et al 2013. Genome of the long‐living sacred lotus (Nelumbo nucifera Gaertn.). Genome Biology 14: R41. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miura K, Hasegawa PM. 2010. Sumoylation and other ubiquitin‐like post‐translational modifications in plants. Trends in Cell Biology 20: 223–232. [DOI] [PubMed] [Google Scholar]
- Moghe GD, Hufnagel DE, Tang H, Xiao Y, Dworkin I, Town CD, Conner JK, Shiu SH. 2014. Consequences of whole‐genome triplication as revealed by comparative genomic analyses of the wild radish Raphanus raphanistrum and three other Brassicaceae species. Plant Cell 26: 1925–1937. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mukhopadhyay D, Dasso M. 2007. Modification in reverse: the SUMO proteases. Trends in Biochemical Sciences 32: 286–295. [DOI] [PubMed] [Google Scholar]
- Murat F, Van de Peer Y, Salse J. 2012. Decoding plant and animal genome plasticity from differential paleo‐evolutionary patterns and processes. Genome Biology and Evolution 4: 917–928. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Myburg AA, Grattapaglia D, Tuskan GA, Hellsten U, Hayes RD, Grimwood J, Jenkins J, Lindquist E, Tice H, Bauer D et al 2014. The genome of Eucalyptus grandis . Nature 510: 356–362. [DOI] [PubMed] [Google Scholar]
- Nei M, Rogozin IB, Piontkivska H. 2000. Purifying selection and birth‐and‐death evolution in the ubiquitin gene family. Proceedings of the National Academy of Sciences, USA 97: 10866–10871. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Novatchkova M, Budhiraja R, Coupland G, Eisenhaber F, Bachmair A. 2004. SUMO conjugation in plants. Planta 220: 1–8. [DOI] [PubMed] [Google Scholar]
- Novatchkova M, Tomanov K, Hofmann K, Stuible HP, Bachmair A. 2012. Update on sumoylation: defining core components of the plant SUMO conjugation system by phylogenetic comparison. New Phytologist 195: 23–31. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Park HJ, Kim WY, Park HC, Lee SY, Bohnert HJ, Yun DJ. 2011. SUMO and SUMOylation in plants. Molecules and Cells 32: 305–316. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Radoshevich L, Impens F, Ribet D, Quereda JJ, Nam Tham T, Nahori MA, Bierne H, Dussurget O, Pizarro‐Cerda J, Knobeloch KP et al 2015. ISG15 counteracts Listeria monocytogenes infection. eLife 4: e06848. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saleh A, Withers J, Mohan R, Marques J, Gu Y, Yan S, Zavaliev R, Nomoto M, Tada Y, Dong X. 2015. Posttranslational modifications of the master transcriptional regulator NPR1 enable dynamic but tight control of plant immune responses. Cell Host & Microbe 18: 169–182. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saracco SA, Miller MJ, Kurepa J, Vierstra RD. 2007. Genetic analysis of SUMOylation in Arabidopsis: conjugation of SUMO1 and SUMO2 to nuclear proteins is essential. Plant Physiology 145: 119–134. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Srilunchang KO, Krohn NG, Dresselhaus T. 2010. DiSUMO‐like DSUL is required for nuclei positioning, cell specification and viability during female gametophyte maturation in maize. Development 137: 333–345. [DOI] [PubMed] [Google Scholar]
- Stamatakis A, Hoover P, Rougemont J. 2008. A rapid bootstrap algorithm for the RAxML Web servers. Systematic Biology 57: 758–771. [DOI] [PubMed] [Google Scholar]
- Tang H, Wang X, Bowers JE, Ming R, Alam M, Paterson AH. 2008. Unraveling ancient hexaploidy through multiply‐aligned angiosperm gene maps. Genome Research 18: 1944–1954. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tomanov K, Hardtke C, Budhiraja R, Hermkes R, Coupland G, Bachmair A. 2012. SUMO conjugating enzyme with active site mutation acts as dominant negative inhibitor of SUMO conjugation in Arabidopsis. Journal of Integrated Plant Biology 55: 75–82. [DOI] [PubMed] [Google Scholar]
- Tomanov K, Zeschmann A, Hermkes R, Eifler K, Ziba I, Grieco M, Novatchkova M, Hofmann K, Hesse H, Bachmair A. 2014. Arabidopsis PIAL1 and 2 promote SUMO chain formation as E4‐type SUMO ligases and are involved in stress responses and sulfur metabolism. Plant Cell 26: 4547–4560. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Van de Peer Y, Fawcett JA, Proost S, Sterck L, Vandepoele K. 2009. The flowering world: a tale of duplications. Trends in Plant Science 14: 680–688. [DOI] [PubMed] [Google Scholar]
- Van den Burg HA, Kini RK, Schuurink RC, Takken FLW. 2010. Arabidopsis small ubiquitin‐like modifier paralogs have distinct functions in development and defense. Plant Cell 22: 1998–2016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- van der Veen AG, Ploegh HL. 2012. Ubiquitin‐like proteins. Annual Review of Biochemistry 81: 323–357. [DOI] [PubMed] [Google Scholar]
- Vierstra RD. 2003. The ubiquitin/26S proteasome pathway, the complex last chapter in the life of many plant proteins. Trends in Plant Science 8: 135–142. [DOI] [PubMed] [Google Scholar]
- Vierstra RD. 2012. The expanding universe of ubiquitin and ubiquitin‐like modifiers. Plant Physiology 160: 2–14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vision TJ, Brown DG, Tanksley SD. 2000. The origins of genomic duplications in Arabidopsis. Science 290: 2114–2117. [DOI] [PubMed] [Google Scholar]
- Wang L, Wansleeben C, Zhao S, Miao P, Paschen W, Yang W. 2014. SUMO2 is essential while SUMO3 is dispensable for mouse embryonic development. EMBO Reports 15: 878–885. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Please note: Wiley Blackwell are not responsible for the content or functionality of any supporting information supplied by the authors. Any queries (other than missing material) should be directed to the New Phytologist Central Office.
Fig. S1 Frequency distribution of the mean synonymous substitution value per synonymous site (Ks) for the retained gene duplicates in the AtSUMO1 and AtSUMO2 duplication block.
Fig. S2 Brassicaceae SUMO5 evolved from an ancient Small Ubiquitin‐Like Modifier (SUMO)‐like paralog found in eudicots.
Fig. S3 Brassicaceae SUMO4 originates from an archetype Small Ubiquitin‐Like Modifier (SUMO) that diversified after the split of Brassicaceae and Cleomaceae.
Fig. S4 Maximum likelihood (ML) tree of Brassicaceae SUMO1, SUMO2 and SUMO3 genes, indicating that Aethionema arabicum SUMO2 groups sister to the SUMO2 and SUMO3 clades.
Fig. S5 Gene tree of the individual ubiquitin‐like modifier (UBL) domains of the DiSUMO‐like (DSUL) locus in grasses with gene identifiers and bootstrap support values indicated.
Table S1 List of the different plant genomes used in this study
Table S2 Annotation of the Brassicaceae Small Ubiquitin‐Like Modifier (SUMO) paralogues, including expression details.
Table S3 Gene IDs of the dicot Small Ubiquitin‐Like Modifier/SUMO‐like (SUMO/SUL) genes studied here in the three different genomic regions: SUMO‐γA (AtSUMO4), SUMO‐γB (AtSUMO1/2) and SUL‐γC (AtSUMO5)
Table S4 Summary of the synteny between Arabidopsis lyrata SUMO5 (ID: 16062200; Al4g17860) and the Small Ubiquitin‐Like Modifier (SUMO) and SUMO‐like (SUL) genes of the basal eudicots grape vine (Vitis vinifera) and eucalyptus (Eucalyptus grandis)


