Figure 3.
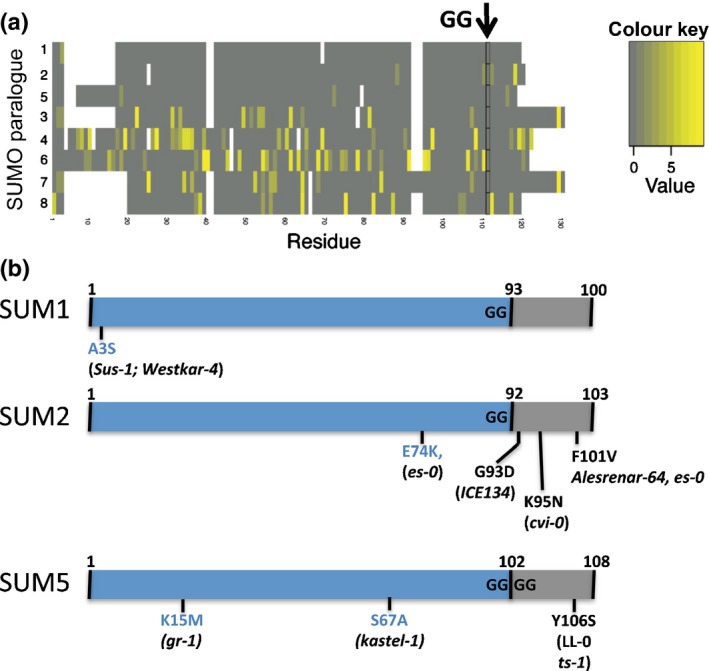
The protein sequences of AtSUMO1, AtSUMO2 and AtSUMO5 are highly conserved in the Arabidopsis population. (a) Heat map diagram of a protein sequence alignment displaying the percentage of amino acid substitutions per residue (grey to yellow) for the different Arabidopsis Small Ubiquitin‐Like Modifier (SUMO) paralogues. AtSUMO1, AtSUMO2 and AtSUMO5 have a few dominant alleles in the population with few accessions carrying an amino acid substitution, whereas, for the other paralogues, many alleles exist in the population with numerous substitutions scattered over the encoded proteins. The diglycine (diGly) motif is indicated (black arrow). The white interruptions indicate gaps in the alignment. (b) Diagram of the three conserved Arabidopsis SUMO paralogues with the substitutions found in the different accessions indicated. Blue, the mature protein; grey, the C‐terminal part removed during processing.
