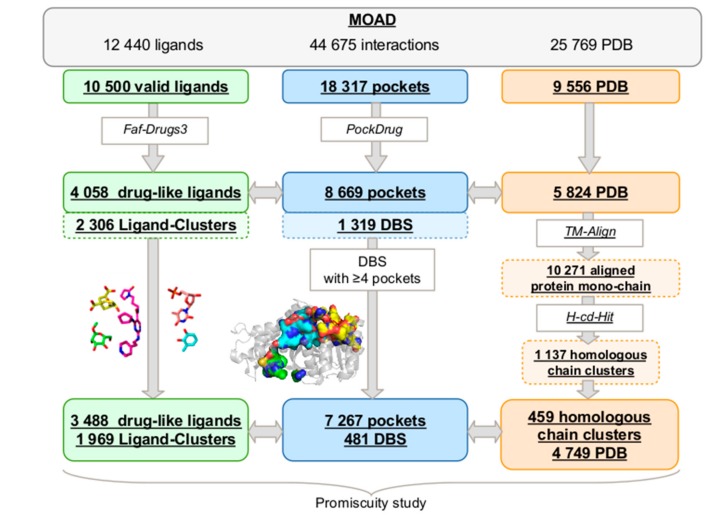
Figure 1.
The protocol developed to extract Druggable Binding Sites (DBS) from the Mother Of All Databases (MOAD) data and to determine their promiscuity. We extracted 3,488 valid and drug-like ligands that were regrouped into 1,969 clusters while using the Tanimoto coefficient (green), 459 homologous chain clusters among the 4,749 Protein Data Bank (PDB) files selected using TM-Align [28] to calculate the identity between all the protein chains, and H-CD-HIT [29] clustering algorithms to group the homologous protein chains (orange). The 7,267 pockets clustered in 481 druggable binding sites (DBS) (blue) correspond to the ligands previously selected and all of the homologous protein chains. The promiscuity was calculated from these data.