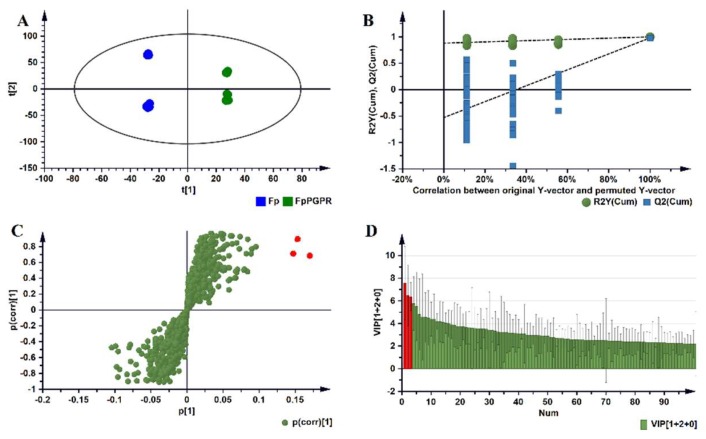
Figure 4.
Orthogonal partial least square-discriminant analysis (OPLS-DA) modelling and variable/feature selection ESI-positive data (roots samples). (A) A typical PCA scores/scatter plot for the OPLS-DA model separating naïve (Fp) versus primed plants (FpPGPR) at 1 d.p.i. (1 + 2 + 0 components, R2X = 0.630, Q2 = 0.980, CV-ANOVA, cross-validated analysis of variance, p-value < 0.05). In the scores plot, it is evident that the two groups are clearly separated: naïve versus primed. (B) A typical response permutation test plot (n = 100) for the OPLS-DA model in (A); the R2 and Q2 values of the permutated models correspond to y-axis intercepts: R2 = (0.0, 8.90) and Q2 = (0.0, 0.542). (C) An OPLS-DA loadings S-plot for the same model in (A); variables situated in the extreme end of the S-plot are statistically relevant and represent leading candidates as discriminating variables/features. (D) Variable importance for the projection (VIP) plot for the same model; pointing mathematically to the importance of each variable (feature) in contributing to group separation in the OPLS-DA model. (C,D) Examples of the variables that were significantly upregulated in primed plants are highlighted in red.