Figure 1.
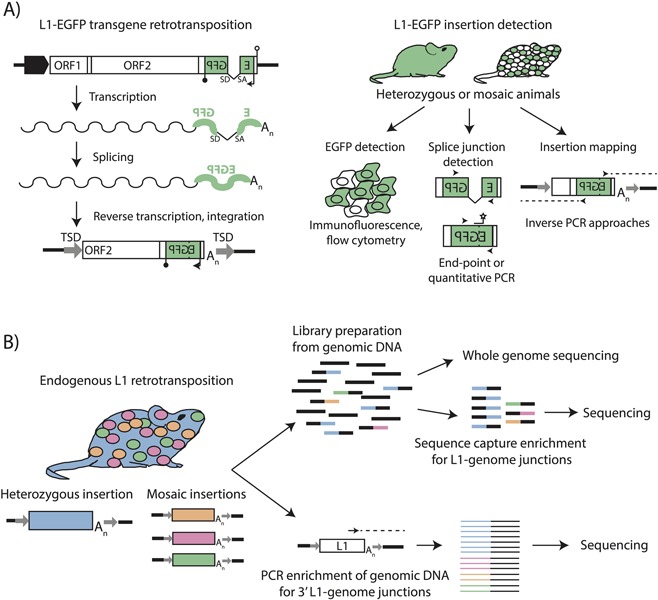
Methods for detecting engineered and endogenous L1 insertions. A) L1‐EGFP transgene retrotransposition. An engineered L1‐EGFP transgene consists of a retrotransposition‐competent L1 element with intact ORFs (white boxes) driven by the native L1 promoter, a heterologous promoter, or both (broad black arrow). The L1 3′UTR contains an enhanced green fluorescent protein (EGFP) reporter gene (green) in antisense orientation to the L1, interrupted by an intron with splice donor (SD) and splice acceptor (SA) sites in sense orientation to the L1. The EGFP reporter is driven by a heterologous promoter (thin black arrow) and terminates in a polyadenylation signal (black lollipop). The EGFP reporter is followed by a polyadenylation signal in sense orientation to the L1 (white lollipop). Upon L1 mRNA transcription (wavy line), the intron in the EGFP gene is spliced. Reverse transcription and integration of the L1 mRNA during target‐primed reverse transcription delivers an intact copy of the EGFP reporter into the genome. B) L1‐EGFP insertion detection. Animals harboring engineered L1‐EGFP insertions can be heterozygous for an insertion (filled green mouse) or mosaic for an insertion (white and green mouse). L1‐EGFP insertions can be detected through expression of the EGFP gene (flow cytometry, fluorescence microscopy), or by PCR‐mediated detection of the spliced EGFP cassette. The genomic locations and structural characteristics of L1‐EGFP insertions can be identified using inverse PCR strategies with primers specific to the EGFP cassette. C) Detection of endogenous L1 retrotransposition. At left, an insertion for which a mouse is consummately heterozygous is represented in blue, and mosaic insertions present at <1 copy per cell are represented in orange, pink, and green. Strategies for detecting endogenous L1 insertions include preparation of Illumina sequencing libraries from genomic DNA (above), which can be directly used for whole genome sequencing (WGS) or enriched for L1‐genome junctions using sequence capture probes, as in RC‐seq. Enrichment for 3′ L1‐genome junctions can also be achieved using PCR‐based approaches (below) in which outward‐facing primers are used to selectively amplify the 3′ termini of active L1 subfamilies, and amplicons subjected to high‐throughput sequencing. In both cases, heterozygous insertions are better represented than mosaic insertions when genomic DNA from bulk tissues is assayed.
