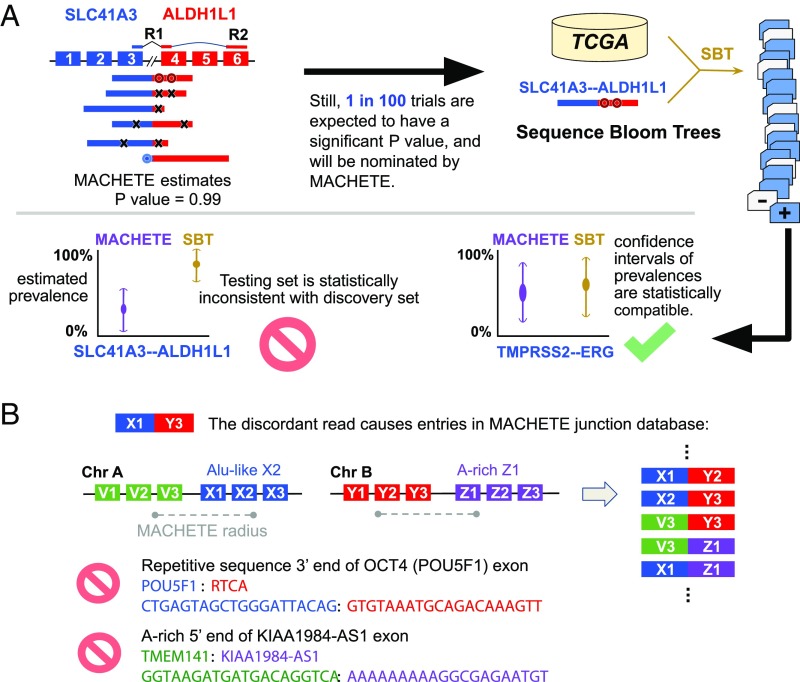
Fig. 1.
Origin and identification of FP from running DEEPEST on thousands of samples. (A) DEEPEST uses all reads, including those censored by other algorithms, to generate an empirical P value for each candidate fusion. SBTs, together with further statistical modeling, are used to identify FP arising from testing on multiple samples, some of which are reported by other algorithms (SI Appendix, Fig. S4A). The first black arrow shows the motivation for designing the SBT step. (B) cDNA or mapping artifacts result in the inclusion of exon–exon junctions from all combinations of exons within a fixed genomic radius of X1 with all exons in the radius of Y3. Some such exon junctions will include degenerate sequences that cannot be mapped uniquely, and thus DEEPEST blinds itself to detection of fusions containing such highly degenerate sequences (for example, due to Alu exonization) or with polyA stretches at the 5′ end.