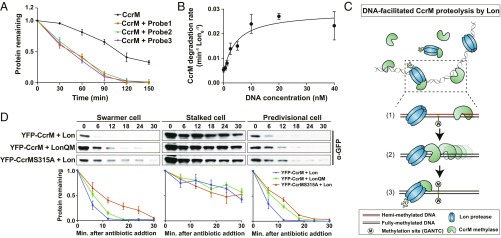
Fig. 3.
DNA serves as a platform in stimulating CcrM proteolysis by Lon. (A) In vitro degradation assays showing the stimulatory effect of DNA on CcrM degradation by Lon. CcrM (1 µM) was incubated with Lon6 (0.25 µM) in the absence or presence of DNA probes (10 nM). The intensity of the CcrM band from 3 independent experiments was quantified. Protein gels are shown in SI Appendix, Fig. S8A. The means ± SDs (n = 3) are plotted. (B) DNA stimulates CcrM degradation by Lon. Degradation rates of CcrM (1 µM) by Lon (0.25 µM) are shown with increasing concentration of DNA probe 1. See Materials and Methods for description of curve fits. (C) A proposed model of DNA-facilitated CcrM degradation by Lon. The Top shows the presence of CcrM, Lon, and DNA fragments in a mixed reaction. A zoomed-in schematic view shows the 3 steps of CcrM degradation by Lon on DNA: first, binding of CcrM and Lon to DNA fragment due to individual high affinity; second, enhanced-intermolecular collision frequency driven by CcrM processivity; third, substrate unfolding and proteolysis. (D) In vivo degradation assays showing the proteolysis of eYFP-CcrM by LonQM and the proteolysis of eYFP-CcrMS315A by Lon in isolated populations of swarmer, stalked, and predivisional (PD) cells in which CcrM and its variant are constitutively expressed. Cells expressing eYFP-CcrM or eYFP- CcrMS315A controlled by Pxyl were grown in M2G and induced with 0.3% xylose, synchronized, and harvested at 0 (swarmer cells), 45 (stalked cells), and 120 (predivisional cells) mps. Samples were treated with chloramphenicol (200 µg/mL) to shut off protein synthesis. Relative protein levels were monitored by immunoblots using anti-CcrM antibody (Top). Band intensities were quantified (Bottom) and error bars represent SDs (n = 4).