Fig. 1. Genetic diversity of E. faecalis strains before and during a hospital outbreak.
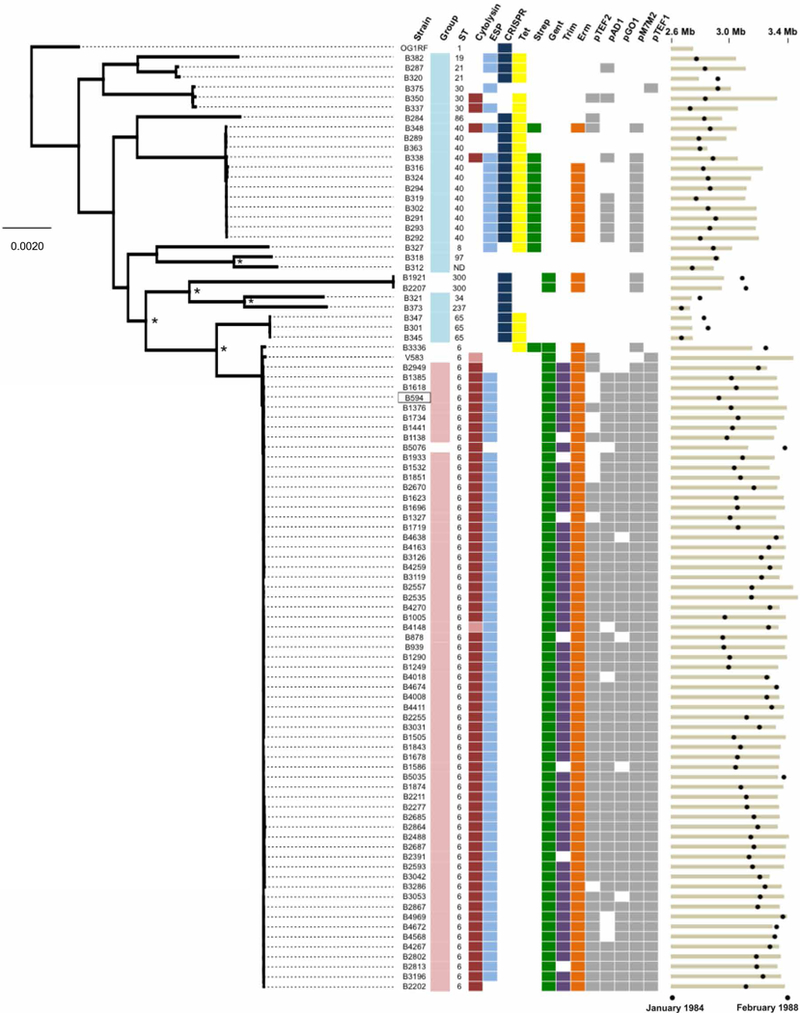
Genetic diversity of E. faecalis strains before (teal) and during (pink) an outbreak from 1984 to 1988 in a Wisconsin hospital is shown. The maximum likelihood phylogeny is shown for the 93 strains sequenced in this study and for the V583 and OG1RF strains (root) that were sequenced previously. The B594 outbreak reference strain is boxed. Orthogroups were clustered with Synergy2, an alignment was generated from single-nucleotide polymorphisms (SNPs) in 1976 single-copy core genes, and a phylogenetic tree was constructed with RAxML. Scale bar shows the number of nucleotide substitutions per site. Asterisks show nodes with bootstrap values of 100; all other nodes have bootstrap values of <90. Colored boxes show the prevalence of virulence (cytolysin and ESP), bacterial immunity (CRISPR-Cas), drug resistance, and plasmid replication genes among pre-outbreak and outbreak E. faecalis strains. Lighter shading in the cytolysin column corresponds to partial, nonfunctional cytolysin operons. Black circles at the far right show the date of isolation of each strain, and tan bars indicate ungapped genome length for each strain. Genome assembly statistics and National Center for Biotechnology Information accession information for all strains are available in table S1.
