Fig. 2. Genetic diversity of E. faecalis outbreak strains.
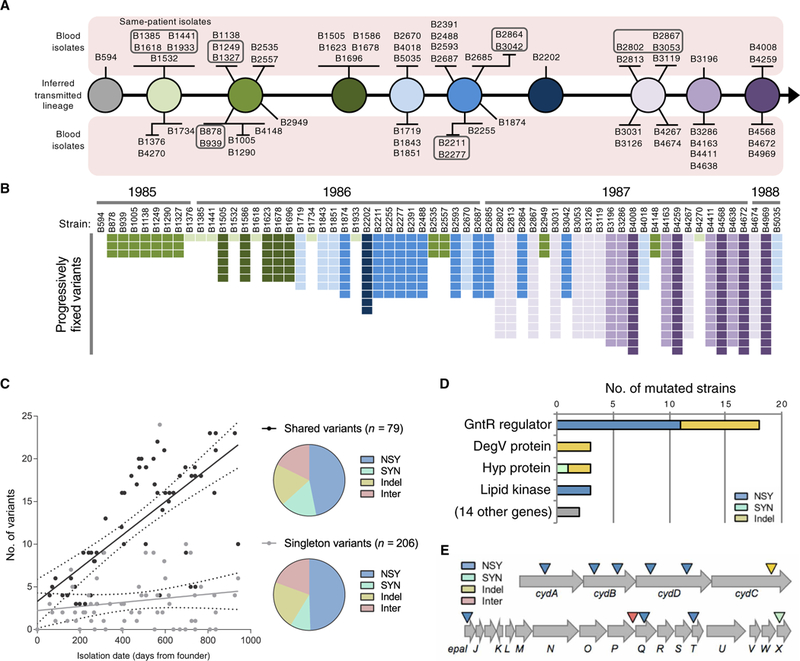
(A) Transmission model of the E. faecalis outbreak lineage. Variants found in 62 outbreak strains were used to infer the genotypes transmitted from patient to patient by the fecal-oral route. The sampled bloodstream isolates are shown as spokes connected to their background genotype and are grouped according to shared variants. Boxed strains were isolated from the same patient. (B) Distribution of progressively fixed variants used to order outbreak strains in (A). Strains are ordered by isolation date, with corresponding years shown on top. The variants are shaded to correspond to the strains in (A). (C) Accumulation of variants over time. Number of shared or singleton variants versus date of isolation is plotted for each strain. NSY, nonsynonymous SNPs; SYN, synonymous SNPs; Indel, insertion/deletion variant; Inter, intergenic variant. (D) Summary of genes that were repeatedly and independently mutated among E. faecalis outbreak strains. Bars show the number of different mutations detected within each gene. (E) Diagram of the cydABDC and epa operons, which were repeatedly and independently mutated among different E. faecalis outbreak strains. Colored triangles indicate the approximate location of mutations.
