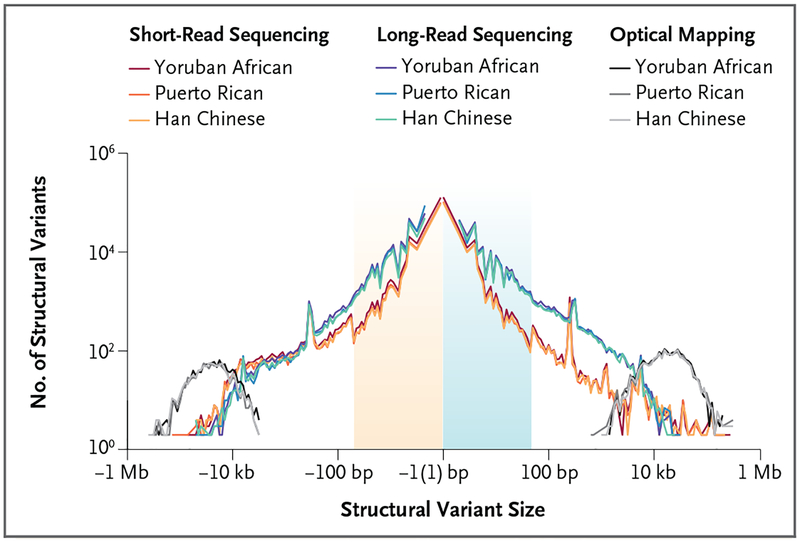
Figure 2. Sensitivity of Structural-Variant Detection with the Use of Different Genomic Technologies.
Shown is a comparison of the number of structural variants (gains or losses of DNA shown on a log scale) as a function of variant size among the same three human genomes, detected with the use of different variant discovery technologies. Structural variants, insertions, and deletions were discovered by short-read sequencing, long-read sequencing (e.g., PacBio), and optical mapping technology (e.g., Bionano Genomics). Short-read sequencing shows reduced sensitivity for the detection of structural variants, especially insertions of 50 bp to 2000 bp. Both technologies underperform for sequence resolution of larger multi-copy-number variants (>10,000 bp); optical mapping technology can be used to detected such variants but not to resolve their sequence organization. Adapted from Chaisson et al.14