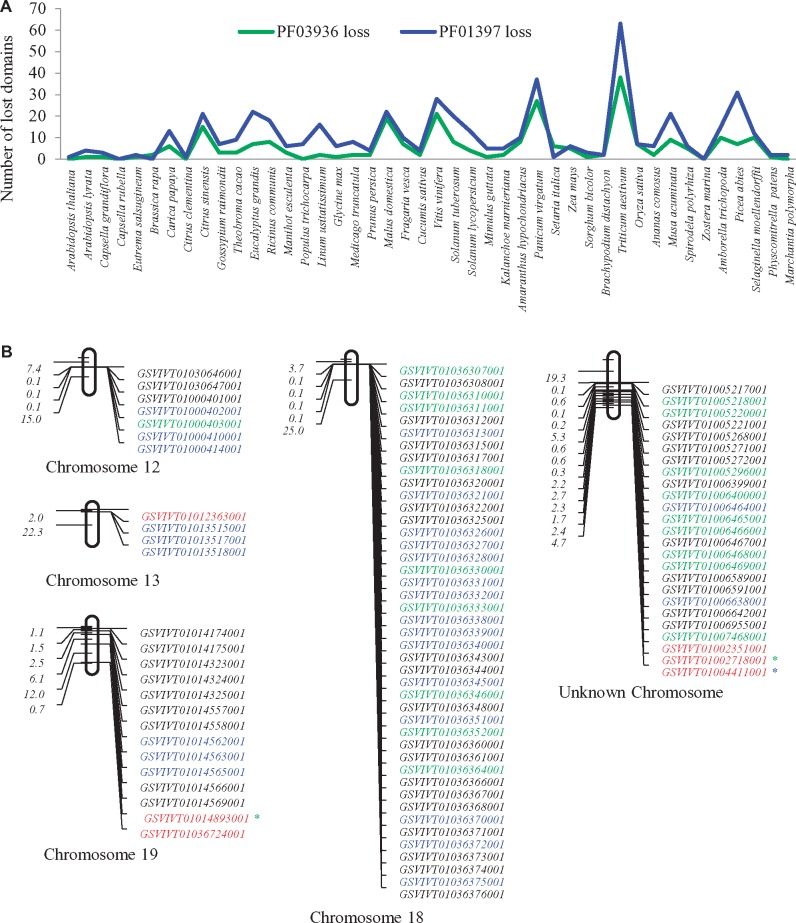
Fig. 6.
—Detection of domain loss and its possible mechanisms. (A) General profiles of both PF01397 and PF03936 domain loss in 44 plant species. (B) Chromosomal distribution of TPSs and detection of tandemly/segmentally duplicated genes in Vitis vinifera. Genes marked with red fonts indicated nontandemly duplicated genes and the remaining genes were all related to tandem duplication. Genes with PF01397 or PF03936 domain loss were labeled with blue or green fonts, respectively. The first value in each chromosome represents the physical position (Mb) of mapped genes. The second value shows the physical distance to the first gene. The rest may be calculated by analogy. Green star “*” indicates that the corresponding gene suffered from PF01397 loss but was not tandemly duplicated gene. The blue star “*” indicates the genes with PF03936 loss that occurred in nontandem duplicated region.