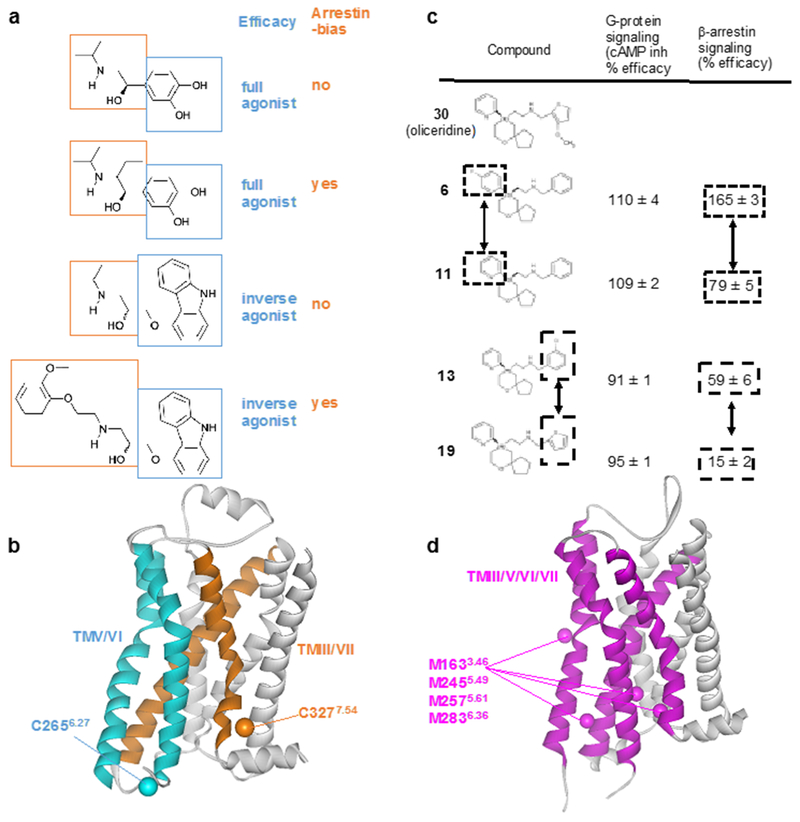
Figure 9 |. NMR screening of biased ligands.
a | Structures and functions of β2AR ligands. Modifications of the ethanolamine tail moieties (highlighted in orange boxes) result in selective modulation of the efficacies for β-arrestin signaling, whereas modifications of the aromatic head groups (highlighted in blue boxes) affect the efficacies of both G protein-mediated signaling and β-arrestin-mediated signaling. b | Crystal structure of the β2AR complex with the inverse agonist carazolol (PDB: 2RH1). TMV and TMVI (cyan) form the binding site for the aromatic head groups shown in (a). TMIII and TMVII (orange) form the binding site for the ethanolamine tail moieties shown in (a). 19F-NMR probes introduced at C2656.27 (Cβ is shown cyan) enable studies of the roles of TMV and TMVI for G protein signaling efficacy. 19F-NMR probes introduced at C3277.54 (Cβ is shown orange) enable studies of TMIII and TMVII in arrestin bias. c | Structure–function relationships of derivatives of the MOR ligand oliceridine. Modification of either of the two aromatic rings that are highlighted in boxes results in selective modulation of β-arrestin signaling. d | Crystal structure of MOR with a morphinan antagonist (PDB: 4DKL). TMIII, TMV, TMVI and TMVII are shown as purple ribbons, and Cε atoms from M1633.46, M2455.49, M2575.61, M2836.36 are shown as purple spheres. NMR studies observing 13CH3 groups of these four methionines suggest that TMIII, TMV, TMVI and TMVII are involved in G protein/arrestin signaling bias.