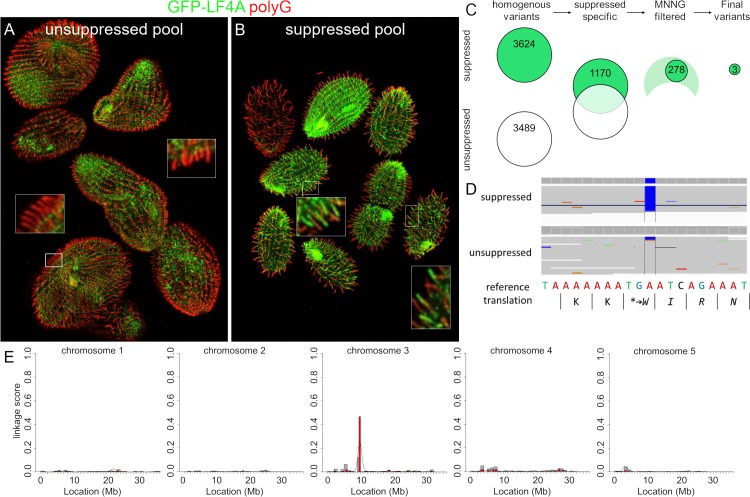
Fig 5. The extragenic suppressor clone SUP1 has a mutation in CDKR1.
(A-B) Immunofluorescence images of (unsuppressed and suppressed) pools of F2 progeny derived from a single sup1/SUP1+ (after an overnight exposure to Cd2+) that were subsequently subjected to whole genome sequencing. Note the patterns of GFP-LF4A inside cilia: the base accumulation in the non-suppressed pool and the ciliary shaft and tip signals in the suppressed pool (see insets for higher magnifications). (C) The results of variant subtraction and filtering based on the alignment of sequencing reads to the macronuclear reference genome. Among the 278 variants consistent with nitrosoguanidine (MNNG) mutagenesis, three candidate variants affect a gene product and have a high fraction of reads supporting the alternative base in the mutant pool (see S1 Table). (D) An IGV browser view of the macronuclear genome sequence of TTHERM_01080590 (CDKR1) that contains the variant scf_8254401:105680 T to C. This point mutation, supported by 100% of the sequencing reads from the mutant pool, changes the stop codon and adds a short peptide to the C-terminus of the predicted product. (E) An allelic composition contrast analysis of the variant co-segregation across all micronuclear chromosomes. The normalized linkage scores show the difference in the allelic composition between the mutant and the wild-type pool at each variant site. This reveals a cluster of variant co-segregation at 9–10 Mb on the micronuclear chromosome 3.