Figure 5.
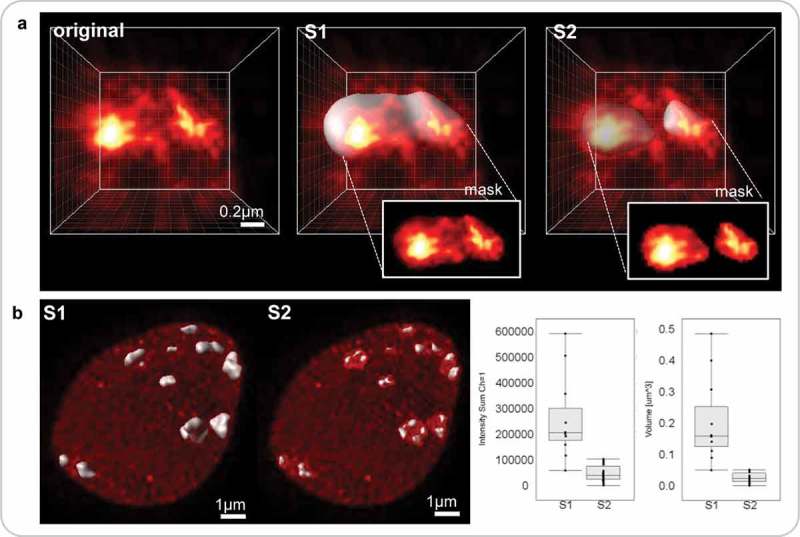
Impact of different segmentation parameters on downstream analyses of nuclear features.
Segmentation parameters influence the shape and boundary of the 3D objects created and will impact downstream analyses in delivering different results of distinct biological significance. Segmentation parameters which may be question-dependent, should thus be defined at the beginning of each batch processing. (a) image detail of a chromocenter from a DNA-stained nucleus imaged by STED and restored by deconvolution (Figure 4A3): original, no segmentation; S1and S2, Imaris-based surface segmentation using the following parameters: background subtraction, largest sphere diameter = 0.4 µm, surface details = 0.1 µm, manual threshold value 24–97 (S1) or 59–166 (S2), without (S1) or with (S2) ‘split touching object’ function (seed point = 0.26 µm). S1 identifies a large, yet heterogeneously staining CC domain while S2 captures individual subdomains in the CC as shown in the insets (channel masks created on each surface). (b) The results from S1 and S2 are shown at the nuclear scale. Both methods yield different results with distinct biological significance (because they capture different object type) regarding the distribution of fluorescence intensity sum and volume of CCs (left and right graphs, respectively). Graphs computed in ImarisVantage.
