ABSTRACT
Background: Intra-tumoral CD8 + T-cell infiltration in squamous cell carcinoma of the head and neck (HNSCC) has previously been linked to the efficacy of cisplatin-based chemoradiation (CDDP-CRTX) and immune checkpoint inhibitor (ICI) monotherapy. Further detailed characterization of the tumor immune-micromilieu and its influence on outcome may guide the development of CRTX-ICI combinations.
Methods: Comprehensive immune transcriptome analysis was applied to a training set of tumor specimens from oropharyngeal squamous cell carcinoma (OPSCC) patients treated with CDDP-CRTX in the ARO-0401 phase III study (n = 33). A composite immune signature risk score (ISRS) for survival prediction was developed, and subsequently validated in two independent OPSCC cohorts treated with either CDDP-CRTX (n = 36) or mitomycin-based CRTX (MMC-CRTX, n = 31). Further validation of the ISRS was performed in the OPSCC subset (n = 79) of the TCGA HNSCC cohort. Potential interference between immune signatures and HPV status was evaluated in multivariate Cox regression models.
Results: Significant differences according to the 3-y OS status in the abundance of tumor-infiltrating T- and B-cells, and the expression levels of 51 immune-related genes were observed. A risk score based on 13 differentially expressed genes involved in cytokine signaling, T-cell effector functions and the TNFR pathway was established as robust predictive factor of OS. Its predictive power was superior to the 6-gene interferon-gamma signature of ICI efficacy and independent of the HPV status.
Conclusions: This study further elucidates the complex interaction of the tumor immune microenvironment with the efficacy of CDDP-CRTX in OPSCC. The results suggest immune markers for selection of patients treated with CRTX-ICI combinations.
KEYWORDS: Immune signature, prediction score, prognostic biomarker, radiochemotherapy, head neck cancer
Introduction
Concurrent chemoradiation (CRTX) is the current standard of care for locally advanced unresectable disease. However, progression-free survival rates at 3 y of less than 40% in the subgroup of HPV-negative HNSCC patients strongly underline the necessity of further treatment optimization. The promising results of immune checkpoint inhibitor (ICI) monotherapy in recurrent/metastatic (R/M) HNSCC have stimulated the clinical development of ICI as a combination partner of CRTX in the curative setting. Recently, exciting results from a phase III clinical trial of CRTX combined with anti-PD-L1 blockade in lung cancer have been reported.1 The rationale for CRTX-ICI combinations is based on accumulating evidence of common immune effector mechanisms linked to the treatment efficacy of both treatment modalities.2 The importance of tumor-infiltrating T-cells for tumor control by radiation was first established in murine models of breast cancer3 and melanoma.4 Subsequent studies of clinical HNSCC cohorts suggested that baseline intra-tumoral T-cell infiltration may not only improve the response likelihood to ICI monotherapies but might also significantly influence the efficacy of CRTX.5,6 In addition, diverse immunogenic properties of platinum-based chemotherapeutics have been described,7 which at least in part are mediated by the induction of costimulatory signals for CD8 + T-cell-dependent tumor destruction.8
We therefore hypothesized that detailed evaluation of the tumor immune microenvironment, going beyond the measurement of single analytes such as CD8 might not only allow the identification of patients who will derive the most benefit from CRTX but might also reveal additional immunologic determinants that could be exploited for rational trial design of CRTX-ICI combinations.
Patients and methods
Patients
Ethical approval for the retrospective biomarker analysis was obtained by the local Ethics Committee (EA2/086/10). Informed consent from all patients was obtained for tissue used in the study. Three independent cohorts of patients with locally advanced oropharyngeal squamous cell carcinoma (OPSCC) who had been treated with concurrent CRTX were included in this immune profiling study. All tumor samples had been collected prior start of definitive CRTX. Archival tumor samples from 64 OPSCC patients who had participated in the prospective randomized multicenter phase III trial ARO-0401 for optimization of concurrent CRTX of stage IVA/B oro-/hypopharyngeal carcinoma9,10 were available for comprehensive immune transcriptome analysis. Thirty-three OPSCC cases from the cisplatin-based CRTX (CDDP-CRTX) arm and 31 OPSCC cases from the mitomycin C-based CRTX (MMC-CRTX) arm of the ARO-0401 trial were included in the training cohort and validation cohort 2, respectively. Validation cohort 1 comprised 36 routine-care OPSCC patients treated with CDDP-CRTX at two German clinical centers (Charité University Hospital Berlin and University Hospital Frankfurt am Main). These cases were selected from the patient cohorts of two previous independent biomarker studies in which the prognostic role of CD8 + T-cell infiltrates5 and the HPV/p16 status (unpublished data) was assessed. The flowchart depicting sample selection in more detail is presented in supplementary Figure 1.
The human papilloma virus (HPV) status was determined by HPV DNA genotyping and p16 immunohistochemistry (IHC) as previously described.11 High-risk HPV DNA positive cases with strong diffuse staining of p16 in >70% of tumor cells were considered HPV-positive.12 Patients’ characteristics are listed in supplementary Table 1 (online only). Kaplan-Meier estimates of OS for training and validation cohorts compared to the remaining ARO-0401 trial population are presented in supplementary Figure 2. Training and validation cohorts were comparable except for a higher proportion of HPV-positive cases in validation cohort 1 which was associated with a significantly improved OS (supplementary Figure 2, c). No significant difference in outcome was observed when only the HPV-negative cases were included in the Kaplan-Meier analysis (supplementary Figure 2, d).
The OPSCC subset of The Cancer Genome Atlas (TCGA) HNSCC provisional dataset was used for further validation (validation cohort 3, supplementary Table 1). RNA sequencing (RNAseq) normalized log2 data (version 1/28/2016), HPV status and clinical data were downloaded from the Broad TCGA GDAC site (http://gdac.broadinstitute.org/).
RNA isolation
FFPE tissue slides (5-µm) were stained with hematoxylin and eosin (HE) and reviewed by a pathologist (W.W. or K.J.) to delineate tumor areas. Only tumor samples with a minimal tumor cell content of 30% were further processed. RNA was isolated from macrodissected tumor areas using the High Pure FFPE RNA Micro Kit (Roche) according to the manufacturer’s protocols. Briefly, tissue sections were deparaffinized with xylene and washed with ethanol. Tissues were lysed and treated with proteinase K for 3 h at 55°C. Thereafter, lysates were applied onto spin columns and after a washing step, total RNA was eluted twice in 40 µl elution buffer. Afterward, the RNA was purified and concentrated using the Clean&Concentrator-5 Kit (Zymo Research, Irvine, CA, USA) according to the manufacturer’s protocol. RNA yield was measured by the Qubit® RNA BR Assay Kit (Thermo Fisher Scientific, Waltham, MA, USA) on the Qubit 3.0 Fluorometer (Thermo Fisher Scientific).
Nanostring analysis
Immune profiles were established using the nCounter PanCancer Immune Profiling Panel and the nCounter Digital Analyzer (nanoString Technologies, Seattle, WA, USA) according to the manufacturer’s protocol. The PanCancer Immune Profiling Panel included 730 immune-related genes and 40 housekeeping genes. Raw expression data were analyzed with the nSolver Analysis Software version 4.0 using the Advanced Analysis Module (nanoString Technologies). For background correction, the geometric mean of negative controls was subtracted from the counts of each gene. The geometric mean of the housekeeping genes plus two times the standard deviation was used to calculate a normalization factor for each sample. Normalized data were log2 transformed for further analysis. Raw and normalized expression data were deposited in Gene Expression Omnibus (GEO) (www.ncbi.nlm.nih.gov/geo/) with accession number GSE113282.
Differently expressed genes (DEGs) were identified in the training cohort by a linear regression model in which a false discovery rate (FDR) of <0.1 determined by the Benjamini-Hochberg algorithm was accepted. Hierarchical clustering analysis of DEGs was performed with the R package “heatmap.plus” (version 3.0.1.) using Euclidean distance and the Ward’s minimum variance method (“ward.D2”).
The analysis of the abundance of various immune cell populations was done using the Cell Type Profiling module of the nSolver Analysis Software which is based on the method described by Danaher and coworkers.13 Briefly, cell populations are quantified using marker genes which are expressed stably and specifically in given cell types.
Development of the Immune Signature Risk Score (ISRS)
Using the R “glmnet” package (version 2.0–5) the least absolute shrinkage and selection operator (LASSO)-penalized Cox regression model was applied to select the immune genes with the strongest prognostic value and lowest correlation among each other.14 Leave-one-out cross-validation was used to determine the optimal value for the shrinkage parameter λ. We chose the value of λ for which the partial likelihood deviance is minimized to the mean cross-validated error plus one standard error. The final ISRS was based on the expression of 13 immune-related genes and their regression coefficients established by the analysis of the training set. The equation for calculation of ISRS-13 is given in the supplementary information (online only). Patient stratification according to ISRS-13 was done using the optimal cut-off value established in the training cohort by receiver operator characteristics (ROC) analysis. Both the model and the cut-off value derived from the training dataset were also applied to the expression data of the validation cohorts 1 and2.
Statistical analysis
For the analysis of the association between immune signatures and outcome the endpoints OS and locoregional control were used, in which an event was defined as death due to any cause (OS) or tumor recurrence at the primary site or regional lymph nodes (LRC), respectively. Event times were measured from the date of start of treatment to the date of event occurrence, censored by the date of the last contact (OS) or the date of the last post-treatment assessment by imaging or clinical examination (LRC). OS curves were plotted using the Kaplan–Meier method and compared between groups with the log-rank test. Uni- and multivariate analyses were performed by the Cox proportional hazard regression. Receiver operating characteristic (ROC) analysis was conducted to evaluate the accuracy of the distinct immune signature scores in predicting the 3-yOS status. Two-tailed Pearson χ2 test was used to test for significant differences of categorical variables and Mann Whitney U test for numerical variables. For all analyses, two-sided P value < .05 was considered statistically significant. Statistical analyses were performed using SPSS version 25.0 (IBM, Armonk, NY, USA).
Results
The tumor immune micromilieu interferes with survival after CRTX
The training cohort was used for assessment of potential differences in the abundance of distinct immune cell subsets in relation to the 3-y OS status. This analysis revealed an increase in tumor-infiltrating lymphocytes (TIL) in pretreatment tumor samples from patients alive compared to those deceased after 3 y(Figure 1, a). Within the lymphocytic infiltrate, the relative portions of B-cells as well as the relative numbers of CD3 + T-cells and their CD8+, cytotoxic and regulatory subsets (Figure 1, b-f) were increased. Regression analysis revealed positive correlations between CD8 + T-cells and cytotoxic (R2 = 0.57; P < .001) as well as regulatory T-cell subsets (R2 = 0.42; P < .001). In contrast, the extent of tumor infiltration by NK cells, macrophages or neutrophils did not differ according to the survival status (Figure 1, g-i).
Figure 1.
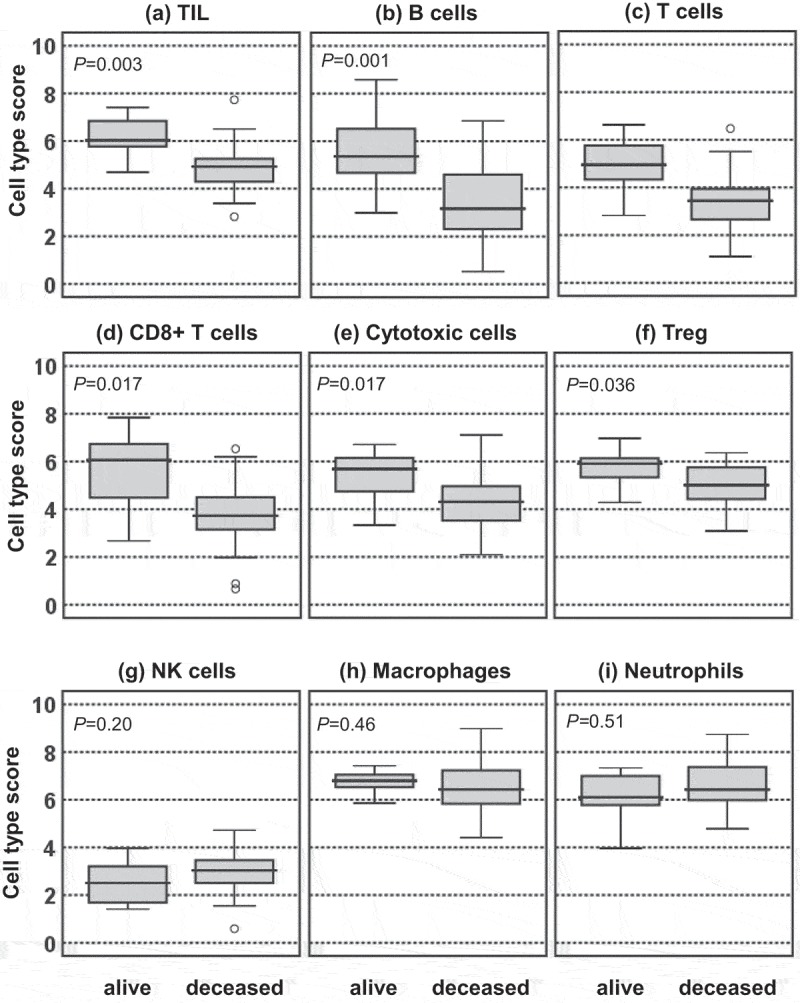
Tumor-immune cell infiltration patterns according to the 3-y OS status. Cell type scores reflecting the abundance of the indicated tumor-infiltrating immune cell types of patients alive or deceased at 3 yafter CRTX are presented in box plots.
Using a false discovery rate of <0.1 in the explorative analysis of the nanoString dataset from the training cohort, we identified 51 immune-related genes as differentially expressed, of which 33 genes were found to be upregulated and 18 genes downregulated in patients alive at 3 yafter start of CRTX (supplementary Figure 3 & supplementary Table 2). Pathway analysis revealed an enrichment of genes from both the innate and adaptive immune system (data not shown). Members of the TNFR superfamily mediating NF-kappaB activation and genes involved in chemoattraction, cytokine signaling as well as T-cell and B-cell receptor signaling were among the 51 DEGs. Hierarchical clustering based on the expression levels of the 51 immune genes (Figure 2, a) revealed two patient subgroups of the training cohort with significantly different OS after CRTX (Figure 2, b).
Figure 2.
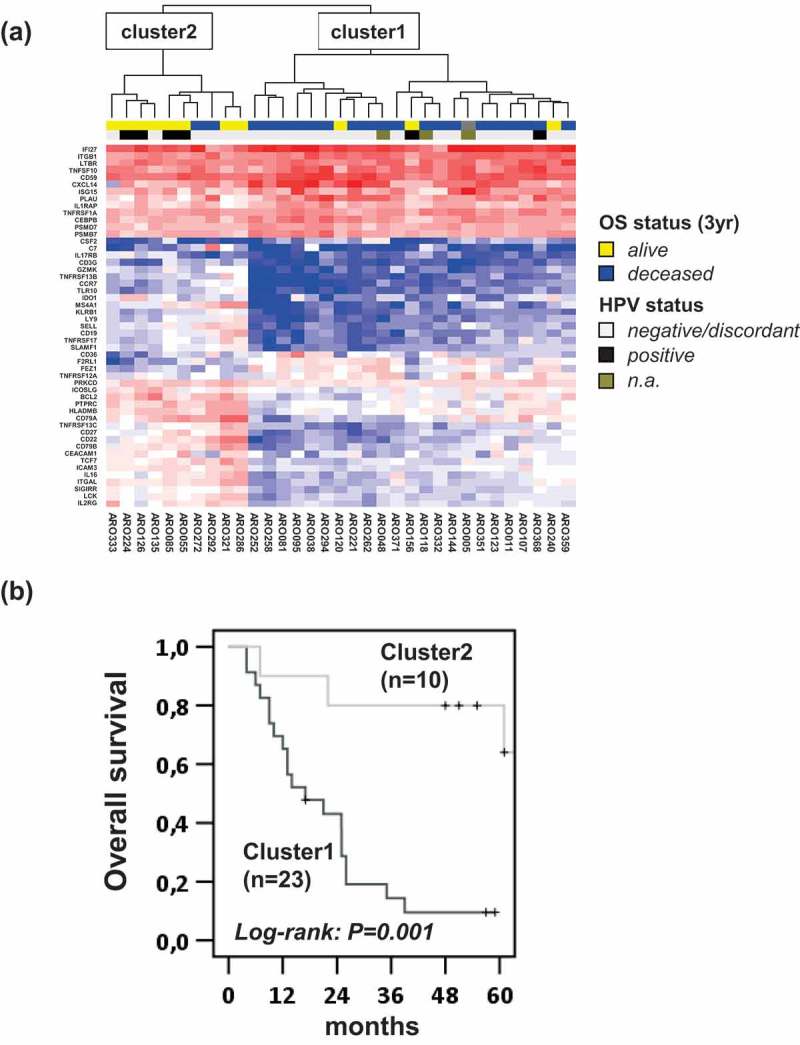
Association of immune expression profiles of tumors and outcome after CDDP-CRTX. (a) Expression levels of 51 differentially expressed immune genes between OPSCC patients alive or dead at 3 yafter the start of CDDP-CRTX were subjected to hierarchical clustering. The 3-y OS and HPV status are depicted in the upper lanes by colored boxes as indicated. (b) Kaplan–Meier curves for OS of patients assigned to cluster 1 and 2 by the hierarchical cluster analysis are shown and log-rank P values are given.
To further assess the prognostic role of the identified candidate immune genes we interrogated the publicly available resource for PREdiction of Clinical Outcomes from Genomic profiles (PRECOG, http://precog.stanford.edu) in which gene expression and clinical outcome data from ~18,000 human tumors across 39 malignancies are integrated.15 Of note, 34 of the 51 immune-related genes (66%) which were differentially expressed according to the 3-yOS status in our study were found to be globally associated with survival in the PRECOG dataset (supplementary Table 2).
Establishment and validation of an immune signature risk score
Using a least absolute shrinkage and selection operator (LASSO)-penalized Cox regression model, a subset of variables was selected from the 51 DEGs in order to establish an immune signature risk score (ISRS) for patient stratification and prognosis in clinical routine. Among the 51 DEGs, 13 genes were identified that showed the highest prognostic value with lowest correlation between each other. An ISRS was calculated as the sum of the weighted log2 expression values of the 13 genes (ISRS-13), using the coefficients established in the LASSO Cox model as weighting factors. We then assessed the accuracy of ISRS-13 in predicting the 3-y OS status using receiver operating characteristic (ROC) analysis. An area under the curve (AUC) of 0.93 (P< .001) confirmed the high accuracy of ISRS-13 in the training cohort (supplementary Figure 4). ROC analysis was also used to define the optimal cutoff for patient stratification. It revealed a sensitivity of 100% and a specificity of 92% for the cut-off value of 18.0. The prognostic value of ISRS-13 was superior to risk classification by the CD8 T-cell score which is based on CD8a and CD8b mRNA expression levels only (supplementary Figure 4).
We then validated the prognostic value of ISRS-13 in three independent patient cohorts. Validation cohort 1 comprised routine-care OPSCC patients treated with CDDP-based CRTX. In order to assess the predictive versus prognostic power of ISRS-13 we also included patients from the ARO-0401 trial who had been treated with MMC-CRTX (validation cohort 2). Tumor samples from the two cohorts were also profiled by the nanostring PanCancer Immune Profiling Panel on the nCounter platform. Algorithms for raw data processing, normalization, calculation of ISRS-13 and the cut-off value for patient stratification were identical to the training cohort. Validation cohort 3 comprised the subset of OPSCC patients from the TCGA HNSCC cohort. Here, RNAseq normalized log2 data were used for calculation of the ISRS-13 and its mean was applied for patient stratification. Kaplan-Meier analysis confirmed the prognostic impact of ISRS-13 on overall survival in all three validation cohorts (Figure 3, b-d). In contrast, a significant influence of ISRS-13 on locoregional tumor control was only observed in the patient group treated with CDDP – but not MMC-based CRTX (Figure 4).
Figure 3.
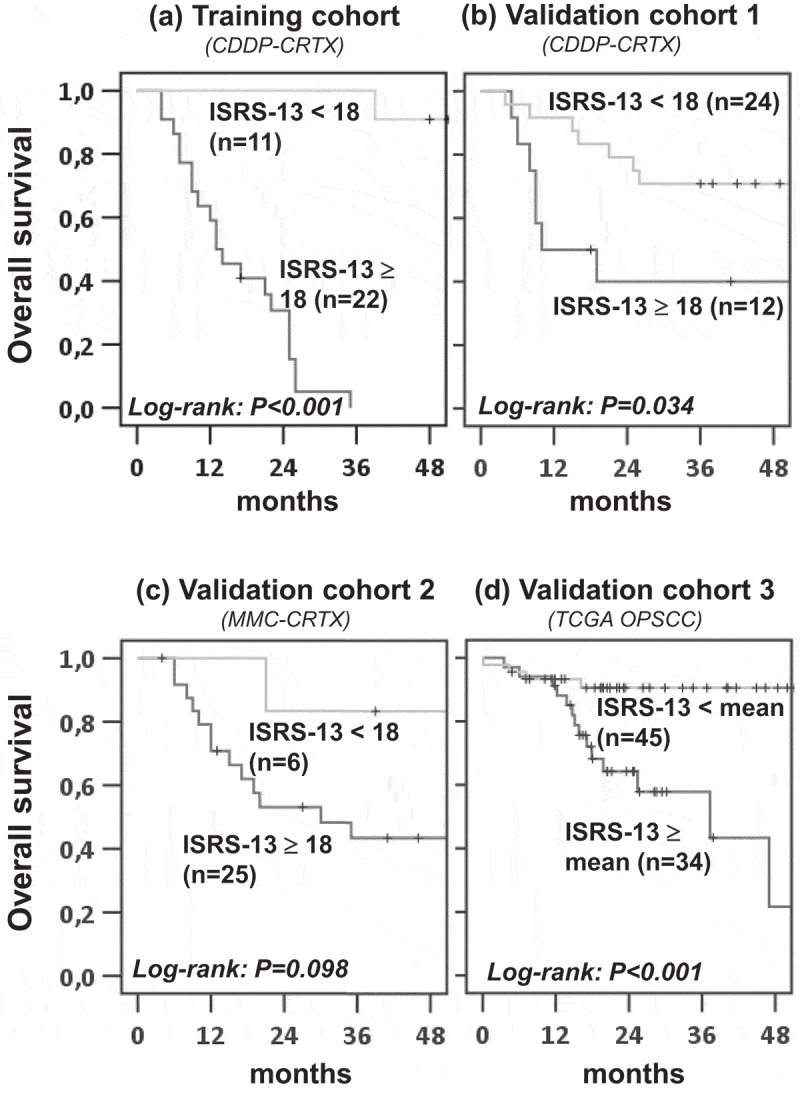
ISRS-13 as robust prognostic factor for overall survival in OPSCC. Patients from the training and validation cohorts were stratified according to ISRS-13 using a cut-off of 18.0 (training, validation cohorts 1 and2) or the mean value (validation cohort 3). Kaplan–Meier curves for OS are shown and log-rank P values are given.
Figure 4.
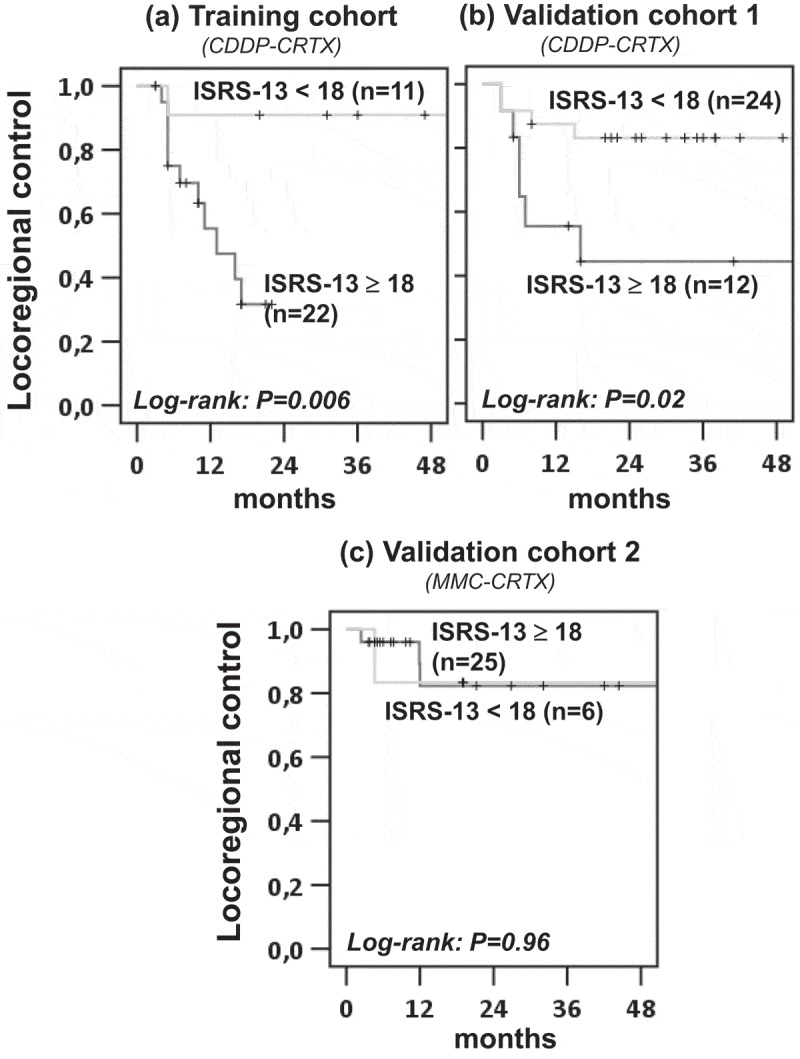
ISRS-13 identifies patients with improved locoregional control from CDDP-CRTX. Patients from the training and validation cohorts were stratified according to ISRS-13 using a cut-off of 18.0. Kaplan–Meier curves for locoregional failure are shown, and log-rank P values are given.
Interference of HPV status and ISRS-13 with overall survival
Given the strong prognostic impact of the HPV status on overall survival after CRTX in OPSCC12 and the previously reported significant differences in the tumor immune micromilieu of HPV-positive and HPV-negative carcinomas,16,17 we evaluated the interaction of both parameters. For this analysis, we pooled the nanoString datasets of the training and validation cohorts since the low prevalence of HPV-positive cases did not provide sufficient statistical power for separate analysis. Overall, HPV-positive tumors, when compared to HPV-negative tumors, displayed significantly increased infiltration of the immune cell subsets (supplementary Figure 5) which were associated with prolonged OS. As expected, we observed a significant accumulation of HPV-positive cases in the patient subgroup with favorable ISRS-13 (i.e. a score <18.0). Nonetheless, patient stratification according to ISRS-13 identified two patient subgroups with significantly different OS in both the HPV-negative and -positive subgroups (Figure 5, a-b).
Figure 5.
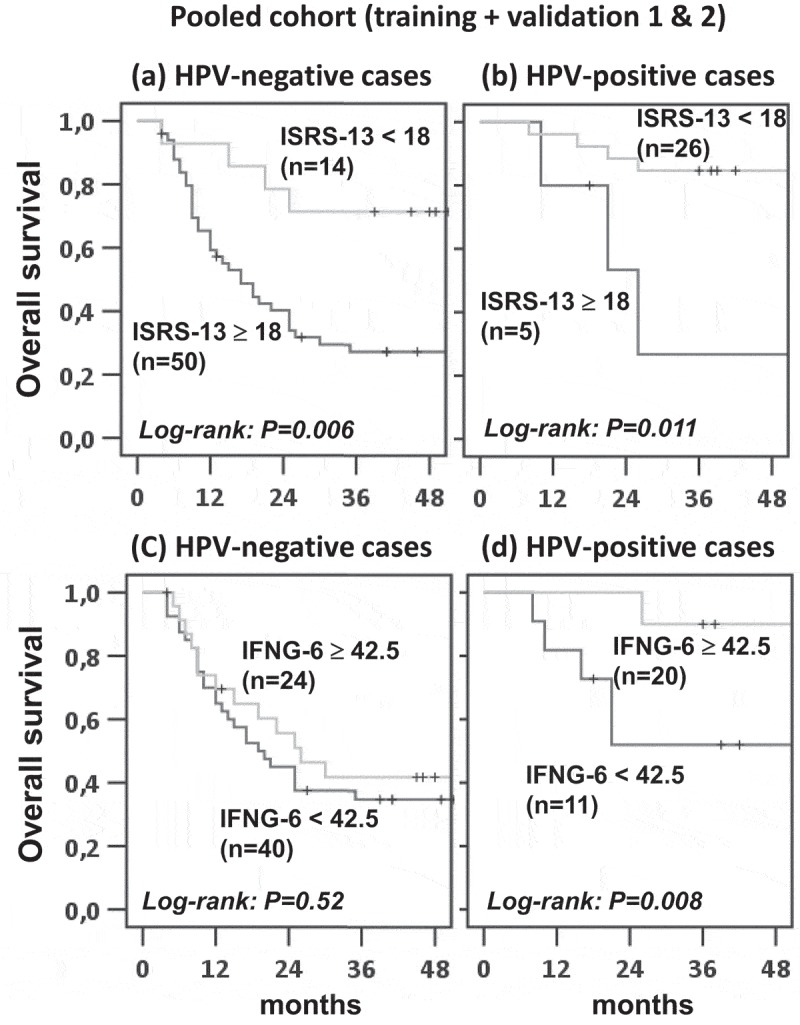
Kaplan-Meier estimates of overall survival among HPV-negative and HPV-positive OPSCC patients, according to the ISRS-13 and IFNG-6 signatures. The HPV-negative and -positive subgroups of patients were stratified according to ISRS-13 (a, b) or IFNG-6 (c, d). Kaplan–Meier curves for OS are shown and log-rank P values are given.
We also evaluated the prognostic value of the 6-gene interferon-gamma (IFNG-6) signature previously reported to discriminate between responders and non-responders to PD-1 blockade in R/M HNSCC18 and other types of cancers.19 Again, the score value with highest specificity/sensitivity identified by ROC analysis (supplementary Figure 4) was used as cut-off (<42.5). While a high IFNG-6 score was significantly associated with improved OS after CRTX in the HPV-positive subgroup of patients, such correlation was absent in the HPV-negative OPSCC population. The significant interaction between the immune signatures and the HPV status in their effects on overall survival was confirmed in the Cox regression model (Table 1). ISRS-13 remained an independent prognosticator for OS in the multivariate model after correcting for tumor stage (Table 1).
Table 1.
Hazard ratios for overall survival, according to the patient group.
| Parameter | Hazard ratio | 95% CI | P value |
|---|---|---|---|
| ISRS-13 | |||
| <18 (reference) | 1.0 | ||
| ≥18 | 2.9 | 1.2-6.8 | 0.015 |
| IFNG-6 | |||
| high (reference) | 1.0 | ||
| low | 1.7 | 0.9-3.2 | 0.105 |
| HPV status | |||
| positive (reference) | 1.0 | ||
| negative | 1.1 | 0.4-3.0 | 0.800 |
| Stage (AJCC 8th edition) | |||
| Stage II / III (reference) | 1.0 | ||
| Stage IVA / IVB | 2.0 | 0.7-5.9 | 0.211 |
| ISRS-13 / p16 IHC status interaction$ | 0.004 | ||
| IFNG-6 / p16 IHC status interaction$ | 0.001 | ||
* For categorization into never/light and heavy smokers a cut-off of >10 packyears was used. $Cox-model, Wald test
Discussion
Beside intrinsic molecular features of tumors significantly interfering with the efficacy of radiation,20–22 the important role of TIL in the outcome of HNSCC patients has been recognized since the early 1980s. Despite limitations in sample size and the inclusion of heterogeneous patient populations in most of the previous studies, previous studies showed with remarkable consistency that tumor-infiltrating T-cells represent a significant independent prognostic variable.23–26 Based on the results of functional studies in preclinical models of melanoma4 and breast cancer,3 it was proposed that the favorable effect of tumor-infiltrating CD8 + T-cells in patients treated with radiotherapy (RTX) relies on a RTX-induced increase in T-cell priming in draining lymphoid tissues, leading to tumor eradication in a CD8 + T cell-dependent fashion.3,4 This hypothesis was further supported by studies of patient cohorts uniformly treated with adjuvant or definitive CRTX in which high infiltration of CD8 + T-cells in tumor tissue before start of treatment was identified as an independent factor of favorable outcome.5,6 Subsequent studies extending the immunohistochemical analysis to PD-L1 suggested additional prognostic value of an expanded immuno-profiling approach.27,28 Here, by comprehensive immune transcriptome analysis we identified further immune-cell subsets and associated immune effector pathways which significantly interfere with outcome after CRTX.
While the importance of CD8 + T-cells, the major killer T-cells for tumor eradication by CRTX is rather well established, it remains less clear why higher infiltrates of Tregs comprising the major immunosuppressive T-cell subset should be beneficial for clinical outcome.29 The positive association of Tregs with survival observed in our study which has already previously been described in head and neck30 and esophageal cancers31 may be due to the known relationship between Tregs and an ‘‘inflamed” tumor microenvironment. As part of a physiological feedback loop preventing immune cell over-activation, Tregs are usually enriched in a TH1 environment. This explains why elevated expression levels of multiple immunosuppressive markers, such as indoleamine-2,3-dioxygenase (IDO) and FOXP3, have been associated with T-cell-inflamed tumors.29 Moreover, in a mouse model of 4-nitroquinoline 1-oxide-induced oral carcinogenesis an increase in inflammatory TH1 immune populations associated with a beneficial antitumor response in tumor-bearing mice was accompanied by an increase in immunosuppressive Tregs in affected lymph nodes.32 Thus, increased infiltration of Tregs in tumors may suggest a background of high TH1 effector response that promotes a favorable outcome. In support of this hypothesis, a strong positive correlation of tumor-infiltrating Tregs and the presence of immune effector cells displaying elevated gene expression of cytolytic enzymes was also observed in our study.
B-cells belong to the poorly characterized TIL subsets in HNSCC and other cancer entities. Only two small cohort studies applying CD20 IHC for detection of tumor-infiltrating B-cells have addressed their prognostic value in HNSCC so far. The first study including a cohort of 33 patients with oro-/hypopharyngeal carcinoma revealed an association of B-cells residing in the peritumoral tissue of lymph node metastases with favorable outcome whereas intraepithelial B-cells in the primary tumor tissue lacked prognostic impact.33 A lack of any prognostic value of tumor-infiltrating B-cells was also reported by a more recent study.34 The discrepancy in the prognostic value of B-cells between our and previous studies might result from different B-cell detection method. Detection based on mRNA expression levels of four B-cell specific genes (CD19, MS4A1, BLK, TNFRSF17) by the nanoString platform might reveal a distinct subpopulation of B-cells than sole IHC staining of CD20, the protein encoded by the MS4A1 gene. This explanation seems very likely in the light of the fact that, similar to T-cells, B-cells represent a heterogeneous population with functionally distinct subsets, and the balance among subtypes can significantly impact on the strength of anti-tumor immunity.35 Beyond presenting antigens, secreting immunoglobulin and directly killing tumor cells, B-cells also produce cytokines that regulate T-cells to inhibit the anti-tumor immune response.35 Clearly, more detailed studies will be necessary to unravel the interference of B-cells with treatment efficacy and tumor progression in HNSCC.
We further demonstrated that 51 of the 730 immune-related genes (7%) were differentially expressed in 3-y survivors compared to non-survivors. The spectrum of the genes associated with the efficacy of CRTX suggests a complex biology of immune cell homeostasis in CRTX-responsive tumors. Their influence in overall survival after CRTX seems not to be unique for OPSCC but to have a more global relevance in cancer since two-thirds of the genes were similarly associated with survival in the pan-cancer PRECOG dataset. Immune profiling based on the 51 DEGs might thus represent a useful tool for patient risk assessment in a larger spectrum of cancer diseases. For routine application, however, a risk score like ISRS-13 which is based on a smaller set of genes might be more practicable. Similar to the previous observation that extended immune signatures were more suitable for capturing the complexity of the dynamic immune response to a tumor,19 the performance of ISRS-13 was superior to sole assessment of CD8a/b expression levels as surrogate marker of CD8 + T-cell infiltrates. ISRS-13 also clearly outperformed the IFNG-6 score in identifying patients with low/high clinical benefit from CRTX, suggesting that the immune cell features contributing to the efficacy of CRTX and ICI therapy only partially overlap. Intriguingly, ISRS-13 was significantly associated with locoregional tumor control by CDDP – but not MMC-based CRTX. This observation is in line with accumulating evidence of significant immunogenic properties of platinum-based chemotherapeutics7,8,36 which have not been reported for MMC. This result strongly support currently pursued strategies of combining CDDP-CRTX with immunomodulatory agents like PD-1 blocking antibodies.37
Previously, the NanoString nCounter platform which we applied for in-depth immune-cell related expression profiling has been extensively used for comprehensive analysis of immune-related genes in archival FFPE tumor tissue.19,38,39 Based on this platform, clinical-grade immune signature assays have been developed which allow prediction of patient response to immune checkpoint inhibitors with high accuracy. Despite any further improvement after initial establishment, performance of immune signatures such as the 18-gene T cell-inflamed gene expression profile was highly robust when applied to independent clinical cohorts,19,39 and compared favorably to IHC analysis.19 Finally yet importantly, gene expression profiling on the NanoString nCounter platform was also possible for very limited amount of biomaterial. Despite these advantageous features of the NanoString nCounter platform, further development of ISRS-13 and its implementation as a clinical-grade diagnostic tool will require validation of gene expression by IHC.
In conclusion, our study further elucidated the complex interaction of the tumor immune microenvironment with the efficacy of CRTX in HPV-positive and HPV-negative OPSCC. The major strength of our study is the analysis of the prognostic value of the tumor immune micromilieu in a uniformly treated OPSCC patient cohort from a phase III clinical trial and the independent validation of our findings in three independent datasets. Limitations are the retrospective nature of the study and the small number of cases in the training and validation cohorts. Further comprehensive immune profiling studies in randomized clinical trials will be necessary for future optimal exploitation of immune effector mechanisms for curative treatment strategies in HNSCC. In addition, the value of ISRS-13 for outcome prediction and the selection of patients with potential preferential benefit from CRTX-ICI combinations will need to be rigorously evaluated in prospective trials.
Funding Statement
This work has been supported by a grant of the Berliner Krebsgesellschaft (TIFF201611 to I.T. and K.J.) and the German Cancer Aid (DKH70-3103 to V.B.; DKH110217 to I.T. and W.W.). The funding sources had no role in the design of this study, its execution, analyses or interpretation of the data, nor in the decision to submit results for publication;Deutsche Krebshilfe [110217];Deutsche Krebshilfe [70-3103];Berliner Krebsgesellschaft [TIFF201611];
Abbreviations
- CRTX
chemoradiation
- CDDP
cisplatin
- MMC
mitomycin C
- OPSCC
oropharyngeal squamous cell carcinoma
- HNSCC
head neck squamous cell carcinoma
- ICI
immune checkpoint inhibitor
- ISRS
immune signature risk score
- DEGs
differentially expressed genes
Acknowledgments
The assistance of the radiation oncologists and pathologists from the clinical centers in retrospective FFPE sample collection is gratefully acknowledged. The results shown here are in part based upon data generated by the TCGA Research Network: http://cancergenome.nih.gov/.
Authors’ contributions
AKH, KJ and IT designed the study. AZ, PB, vdGJ, CR, KJ, and WW contributed to sample collection. KJ and WW supervised the generation of FFPE tissue sections and performed their pathological evaluation. AKH processed samples for mRNA analysis by the nanoString platform. AKH and IT performed the statistical analyses. IT, AKH, KJ, AZ, PB, vdGJ, CR, MH, UK, and VB analyzed and discussed the results. IT and AKH jointly developed the structure and arguments for the paper. IT wrote the manuscript. All authors discussed, made critical revisions and approved the final manuscript.
Disclosure of Potential Conflicts of Interest
No potential conflicts of interest were disclosed.
Supplementary material
Supplementary material data can be accessed here
References
- 1.Antonia SJ, Villegas A, Daniel D, Vicente D, Murakami S, Hui R, Yokoi T, Chiappori A, Lee KH, de Wit M, et al. Durvalumab after chemoradiotherapy in stage iii non-small-cell lung cancer. N Engl J Med. 2017;377(20):1919–1929. doi: 10.1056/NEJMoa1709937. [DOI] [PubMed] [Google Scholar]
- 2.Sharabi AB, Lim M, DeWeese TL, Drake CG.. Radiation and checkpoint blockade immunotherapy: radiosensitisation and potential mechanisms of synergy. Lancet Oncol. 2015;16(13):e498–509. doi: 10.1016/S1470-2045(15)00007-8. [DOI] [PubMed] [Google Scholar]
- 3.Demaria S, Kawashima N, Yang AM, Devitt ML, Babb JS, Allison JP, Formenti SC.. Immune-mediated inhibition of metastases after treatment with local radiation and CTLA-4 blockade in a mouse model of breast cancer. Clin Cancer Res. 2005;11:728–734. [PubMed] [Google Scholar]
- 4.Lee Y, Auh SL, Wang Y, Burnette B, Wang Y, Meng Y, Beckett M, Sharma R, Chin R, Tu T, et al. Therapeutic effects of ablative radiation on local tumor require CD8+ T cells: changing strategies for cancer treatment. Blood. 2009;114(3):589–595. doi: 10.1182/blood-2009-02-206870. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Balermpas P, Michel Y, Wagenblast J, Seitz O, Weiss C, Rodel F, Rödel C, Fokas E. Tumour-infiltrating lymphocytes predict response to definitive chemoradiotherapy in head and neck cancer. Br J Cancer. 2014;110(2):501–509. doi: 10.1038/bjc.2013.640. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Balermpas P, Rodel F, Rodel C, Krause M, Linge A, Lohaus F, Baumann M, Tinhofer I, Budach V, Gkika E, et al. CD8+ tumour-infiltrating lymphocytes in relation to HPV status and clinical outcome in patients with head and neck cancer after postoperative chemoradiotherapy: A multicentre study of the German cancer consortium radiation oncology group (DKTK-ROG). Int J Cancer. 2016;138(1):171–181. doi: 10.1002/ijc.29683. [DOI] [PubMed] [Google Scholar]
- 7.Hato SV, Khong A, de Vries IJ, Lesterhuis WJ. Molecular pathways: the immunogenic effects of platinum-based chemotherapeutics. Clin Cancer Res. 2014;20(11):2831–2837. doi: 10.1158/1078-0432.CCR-13-3141. [DOI] [PubMed] [Google Scholar]
- 8.Beyranvand Nejad E, van der Sluis TC, van Duikeren S, Yagita H, Janssen GM, van Veelen PA, Melief CJM, van der Burg SH, Arens R. Tumor eradication by cisplatin is sustained by CD80/86-mediated costimulation of CD8+ T cells. Cancer Res. 2016;76(20):6017–6029. doi: 10.1158/0008-5472.CAN-16-0881. [DOI] [PubMed] [Google Scholar]
- 9.Budach V, Cho CH, Sedlmaier B, Wittlinger M, Iro H, Engenhart-Cabillic R, Hautmann M, Strutz J, Flentje M, Hueltenschmidt B, et al. Five years’ results of the German ARO 04-01 trial of concurrent 72 Gy hyperfractionated accelerated radiation therapy (HART) plus once weekly cisplatinum/5-FU versus mitomycin C/5-FU in stage IV head and neck cancer. J Clin Oncol. 2012;30(suppl):abstr5512. [Google Scholar]
- 10.Hess A-K, Müer A, Mairinger FD, Weichert W, Stenzinger A, Hummel M, Budach V, Tinhofer I. MiR-200b and miR-155 as predictive biomarkers for the efficacy of chemoradiation in locally advanced head and neck squamous cell carcinoma. Eur J Cancer. 2017;77:3–12. doi: 10.1016/j.ejca.2017.02.018. [DOI] [PubMed] [Google Scholar]
- 11.Tinhofer I, Stenzinger A, Eder T, Konschak R, Niehr F, Endris V, Distel L, Hautmann MG, Mandic R, Stromberger C, et al. Targeted next-generation sequencing identifies molecular subgroups in squamous cell carcinoma of the head and neck with distinct outcome after concurrent chemoradiation. Ann Oncol. 2016;27(12):2262–2268. doi: 10.1093/annonc/mdw426. [DOI] [PubMed] [Google Scholar]
- 12.Ang KK, Harris J, Wheeler R, Weber R, Rosenthal DI, Nguyen-Tan PF, Westra WH, Chung CH, Jordan RC, Lu C, et al. Human papillomavirus and survival of patients with oropharyngeal cancer. N Engl J Med. 2010;363(1):24–35. doi: 10.1056/NEJMoa0912217. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Danaher P, Warren S, Dennis L, D’Amico L, White A, Disis ML, Geller MA, Odunsi K, Beechem J, Fling SP. Gene expression markers of tumor infiltrating leukocytes. J Immunother Cancer. 2017;5:18. doi: 10.1186/s40425-017-0215-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Tibshirani R. The lasso method for variable selection in the Cox model. Stat Med. 1997;16:385–395. [DOI] [PubMed] [Google Scholar]
- 15.Gentles AJ, Newman AM, Liu CL, Bratman SV, Feng W, Kim D, Nair VS, Xu Y, Khuong A, Hoang CD, et al. The prognostic landscape of genes and infiltrating immune cells across human cancers. Nat Med. 2015;21(8):938–945. doi: 10.1038/nm.3909. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Keck MK, Zuo Z, Khattri A, Stricker TP, Brown CD, Imanguli M, Rieke D, Endhardt K, Fang P, Brägelmann J, et al. Integrative analysis of head and neck cancer identifies two biologically distinct HPV and three non-HPV subtypes. Clin Cancer Res. 2015;21(4):870–881. doi: 10.1158/1078-0432.CCR-14-2481. [DOI] [PubMed] [Google Scholar]
- 17.Wood O, Woo J, Seumois G, Savelyeva N, McCann KJ, Singh D, Jones T, Peel L, Breen MS, Ward M, et al. Gene expression analysis of TIL rich HPV-driven head and neck tumors reveals a distinct B-cell signature when compared to HPV independent tumors. Oncotarget. 2016;7(35):56781–56797. doi: 10.18632/oncotarget.10788. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Seiwert TY, Burtness B, Mehra R, Weiss J, Berger R, Eder JP, Heath K, McClanahan T, Lunceford J, Gause C, et al. Safety and clinical activity of pembrolizumab for treatment of recurrent or metastatic squamous cell carcinoma of the head and neck (KEYNOTE-012): an open-label, multicentre, phase 1b trial. Lancet Oncol. 2016;17(7):956–965. doi: 10.1016/S1470-2045(16)30066-3. [DOI] [PubMed] [Google Scholar]
- 19.Ayers M, Lunceford J, Nebozhyn M, Murphy E, Loboda A, Kaufman DR, Albright A, Cheng JD, Kang SP, Shankaran V, et al. IFN-γ-related mRNA profile predicts clinical response to PD-1 blockade. J Clin Invest. 2017;127(8):2930–2940. doi: 10.1172/JCI91190. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Foy JP, Bazire L, Ortiz-Cuaran S, Deneuve S, Kielbassa J, Thomas E, Viari A, Puisieux A, Goudot P, Bertolus C, et al. A 13-gene expression-based radioresistance score highlights the heterogeneity in the response to radiation therapy across HPV-negative HNSCC molecular subtypes. BMC Med. 2017;15(1):165. doi: 10.1186/s12916-017-0929-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Horn D, Hess J, Freier K, Hoffmann J, Freudlsperger C. Targeting EGFR-PI3K-AKT-mTOR signaling enhances radiosensitivity in head and neck squamous cell carcinoma. Expert Opin Ther Targets. 2015;19(6):795–805. doi: 10.1517/14728222.2015.1012157. [DOI] [PubMed] [Google Scholar]
- 22.Freudlsperger C, Horn D, Weissfuss S, Weichert W, Weber KJ, Saure D, Sharma S, Dyckhoff G, Grabe N, Plinkert P, et al. Phosphorylation of AKT(Ser473) serves as an independent prognostic marker for radiosensitivity in advanced head and neck squamous cell carcinoma. Int J Cancer. 2015;136(12):2775–2785. doi: 10.1002/ijc.29328. [DOI] [PubMed] [Google Scholar]
- 23.Lei Y, Xie Y, Tan YS, Prince ME, Moyer JS, Nor J, Wolf GT. Telltale tumor infiltrating lymphocytes (TIL) in oral, head & neck cancer. Oral Oncol. 2016;61:159–165. doi: 10.1016/j.oraloncology.2016.08.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Punt S, Dronkers EA, Welters MJ, Goedemans R, Koljenovic S, Bloemena E, Snijders PJF, Gorter A, van der Burg SH, Baatenburg de Jong RJ, et al. A beneficial tumor microenvironment in oropharyngeal squamous cell carcinoma is characterized by a high T cell and low IL-17(+) cell frequency. Cancer Immunol Immunother. 2016;65(4):393–403. doi: 10.1007/s00262-016-1805-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Kogashiwa Y, Yasuda M, Sakurai H, Nakahira M, Sano Y, Gonda K, Ikeda T, Inoue H, Kuba K, Oba S, et al. PD-L1 expression confers better prognosis in locally advanced oral squamous cell carcinoma. Anticancer Res. 2017;37(3):1417–1424. doi: 10.21873/anticanres.11465. [DOI] [PubMed] [Google Scholar]
- 26.Xu Q, Wang C, Yuan X, Feng Z, Han Z. Prognostic value of tumor-infiltrating lymphocytes for patients with head and neck squamous cell carcinoma. Transl Oncol. 2017;10(1):10–16. doi: 10.1016/j.tranon.2016.10.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Balermpas P, Rodel F, Krause M, Linge A, Lohaus F, Baumann M, Tinhofer I, Budach V, Sak A, Stuschke M, et al. The PD-1/PD-L1 axis and human papilloma virus in patients with head and neck cancer after adjuvant chemoradiotherapy: A multicentre study of the German cancer consortium radiation oncology group (DKTK-ROG). Int J Cancer. 2017;141(3):594–603. doi: 10.1002/ijc.30770. [DOI] [PubMed] [Google Scholar]
- 28.Solomon B, Young RJ, Bressel M, Urban D, Hendry S, Thai A, Angel C, Haddad A, Kowanetz M, Fua T, et al. Prognostic significance of PD-L1+ and CD8+ immune cells in HPV+ oropharyngeal squamous cell carcinoma. Cancer Immunol Res. 2018;6:295–304. doi: 10.1158/2326-6066.CIR-17-0299. [DOI] [PubMed] [Google Scholar]
- 29.Spranger S, Spaapen RM, Zha Y, Williams J, Meng Y, Ha TT, Gajewski TF. Up-regulation of PD-L1, IDO, and T(regs) in the melanoma tumor microenvironment is driven by CD8(+) T cells. Sci Transl Med. 2013;5(200):200ra116. doi: 10.1126/scitranslmed.3006504. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Badoual C, Hans S, Rodriguez J, Peyrard S, Klein C, Agueznay Nel H, Mosseri V, Laccourreye O, Bruneval P, Fridman WH, et al. Prognostic value of tumor-infiltrating CD4+ T-cell subpopulations in head and neck cancers. Clin Cancer Res. 2006;12(2):465–472. doi: 10.1158/1078-0432.CCR-05-1886. [DOI] [PubMed] [Google Scholar]
- 31.Shang B, Liu Y, Jiang SJ, Liu Y. Prognostic value of tumor-infiltrating FoxP3+ regulatory T cells in cancers: a systematic review and meta-analysis. Sci Rep. 2015;5:15179. doi: 10.1038/srep15179. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.De Costa AM, Schuyler CA, Walker DD, Young MR. Characterization of the evolution of immune phenotype during the development and progression of squamous cell carcinoma of the head and neck. Cancer Immunol Immunother. 2012;61(6):927–939. doi: 10.1007/s00262-011-1154-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Pretscher D, Distel LV, Grabenbauer GG, Wittlinger M, Buettner M, Niedobitek G. Distribution of immune cells in head and neck cancer: CD8+ T-cells and CD20+ B-cells in metastatic lymph nodes are associated with favourable outcome in patients with oro- and hypopharyngeal carcinoma. BMC Cancer. 2009;9:292. doi: 10.1186/1471-2407-9-292. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Schneider K, Marbaix E, Bouzin C, Hamoir M, Mahy P, Bol V, Grégoire V. Immune cell infiltration in head and neck squamous cell carcinoma and patient outcome: a retrospective study. Acta Oncol. 2018 Sep;57(9):1165–1172. doi: 10.1080/0284186X.2018.1445287. [DOI] [PubMed] [Google Scholar]
- 35.Tsou P, Katayama H, Ostrin EJ, Hanash SM. The emerging role of B cells in tumor immunity. Cancer Res. 2016;76(19):5597–5601. doi: 10.1158/0008-5472.CAN-16-0431. [DOI] [PubMed] [Google Scholar]
- 36.Aranda F, Bloy N, Pesquet J, Petit B, Chaba K, Sauvat A, Kepp O, Khadra N, Enot D, Pfirschke C, et al. Immune-dependent antineoplastic effects of cisplatin plus pyridoxine in non-small-cell lung cancer. Oncogene. 2015;34(23):3053–3062. doi: 10.1038/onc.2014.234. [DOI] [PubMed] [Google Scholar]
- 37.Johnson CB, Win SY, Pullikuth A, Jin G, Su J, Chou JW, Hoadley KA, Print C, Knowlton N, Black MA, et al. Combination therapy with PD-1/PD-L1 blockade: an overview of ongoing clinical trials. Oncoimmunology. 2018;7(4):e1408744. doi: 10.1080/2162402X.2018.1490854. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Ribas A, Robert C, Hodi FS, Wolchok JD, Joshua AM, Hwu W-J, Weber JS, Zarour HM, Kefford R, Loboda A, et al. Association of response to programmed death receptor 1 (PD-1) blockade with pembrolizumab (MK-3475) with an interferon-inflammatory immune gene signature. J Clin Oncol. 2015;33(15_suppl):3001. doi: 10.1200/jco.2015.33.15_suppl.3001. [DOI] [Google Scholar]
- 39.Cristescu R, Mogg R, Ayers M, Albright A, Murphy E, Yearley J, Sher X, Liu XQ, Lu H, Nebozhyn M, et al. Pan-tumor genomic biomarkers for PD-1 checkpoint blockade-based immunotherapy. Science. 2018;362:6411. doi: 10.1126/science.aar3593. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


