Extended Data Figure 1:
Photostimulation characterization and methods
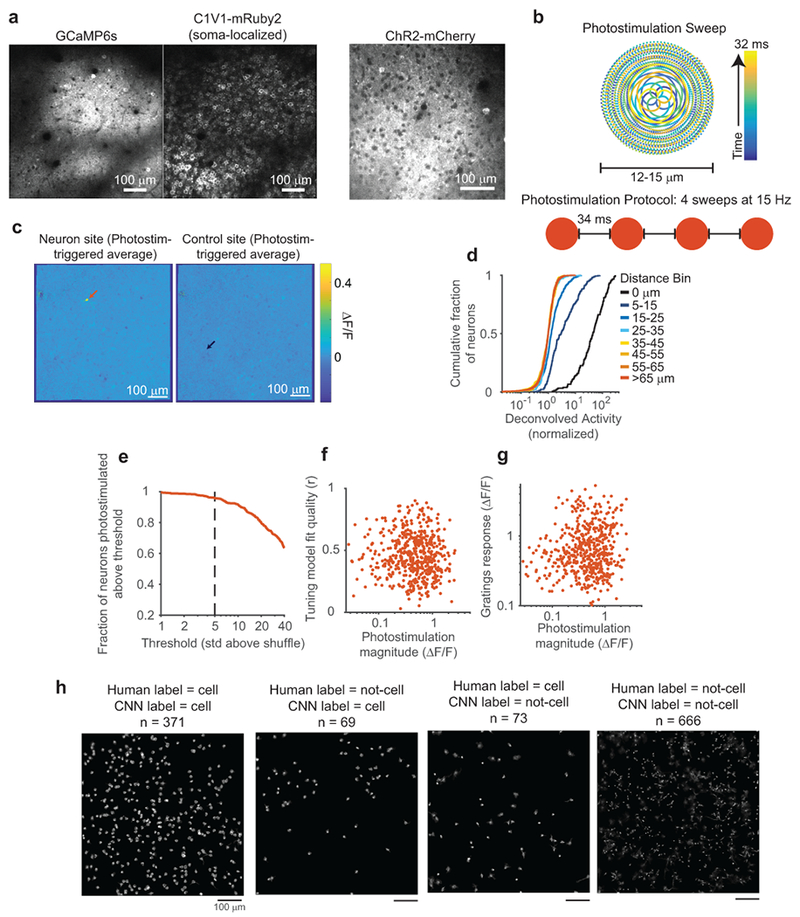
(a) Left, images showing GCaMP6s and densely expressed, soma-localized C1V1 in the same neurons. Right, an image of Channelrhodopsin-2 tagged with mCherry, obtained from a different mouse. Note that non-localized channels are prominent in the neuropil background compared with soma-localized channels.
(b) Photostimulation protocol schematic. Top: beam position as a function of time, samples of mirror trajectory plotted at 100 kHz. Bottom: Four repeats of an identical sweep were used to photostimulate neurons.
(c) Photostimulation triggered average images, for a neuron (left) and control (right) site from the experiment in Fig. 1b. Arrows mark the location of both sites.
(d) Cumulative density plots of photostimulated neuron responses for different lateral displacements of target location from the neuron’s center. Same data as in Fig. 1e, but note log scale of x-axis. The 15-25 μm offset caused responses that were not present at greater distances.
(e) Fraction of neurons that could be photostimulated as a function of the threshold for this classification. At a threshold of 5 std above shuffle, more than 96% of neurons (n=518) could be photostimulated. Shuffle distributions were computed by bootstrap resampling of activity from trials the neuron was not targeted.
(f) Fit quality of the GP tuning model vs. photostimulation magnitude. Each dot is a single targeted neuron (n = 518 neurons). Spearman correlation, c = 0.084, p = 0.055.
(g) Mean gratings response of a neuron vs. photostimulation magnitude. Each dot is a single targeted neuron (n = 518 neurons). Spearman correlation, c = 0.11, p = 0.009.
(h) A CNN was trained with human-labeled data to predict whether CNMF sources were identified as a cell body or an alternative source, including distinct neural processes, excessively blurry or out-of-plane cells, or artefactual sources (see Methods). Note that many non-soma sources exhibited similar calcium transient signals as cell body sources. Because there is no objective ground-truth for this classification, held-out datasets were hand labeled, and compared to CNN labeling. One example dataset is shown here. The large majority of sources were labeled identically, however there are borderline cases where labels differed; many cases appear to be either human error in labeling, due to finite human time and inconsistencies in making borderline judgments, or an overly conservative CNN criteria for cell classification. Neither of these errors are expected to impact results presented in this manuscript.
