Figure 1.
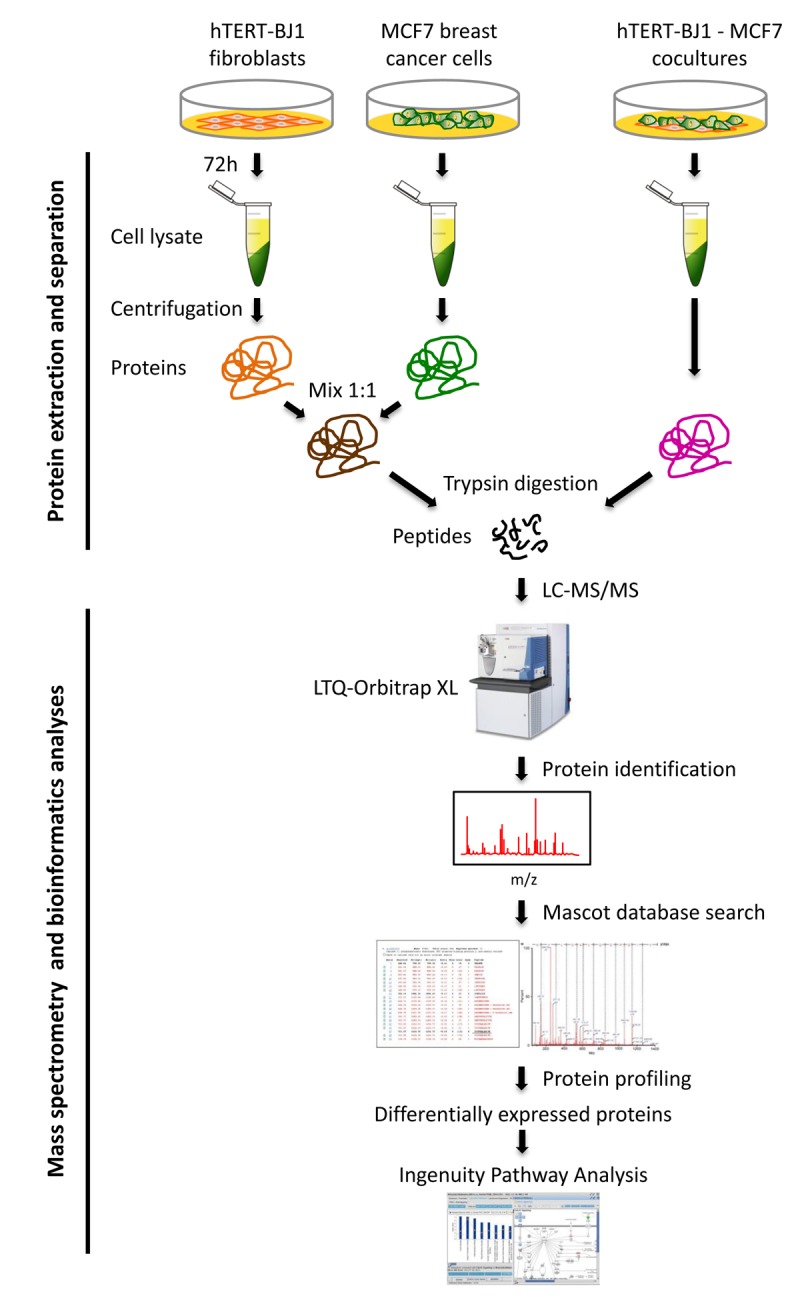
Schematic diagram summarizing the work‐flow for MCF7‐fibroblast co‐culture studies and bioinformatics validation. Protein lysates were obtained from hTERT‐BJ1 fibroblasts after 72 h co‐culture with MCF7 breast cancer cells. Alternatively, protein lysates were obtained from hTERT‐BJ1 fibroblasts and MCF7 cells cultured separately as monolayers and then mixed. Peptides obtained after trypsin digestion were analysed via LC‐MS/MS on an LTQ‐ Orbitrap XL mass spectrometer. Label‐free quantitative proteomics was used to detect changes in protein abundances across co‐cultures and mixed cell population extracts. The proteomics data sets were further analyzed using Ingenuity Pathway Analysis. This co‐culture approach is predicted to better simulate the fibroblast‐rich local tumor micro‐environment in vivo.
