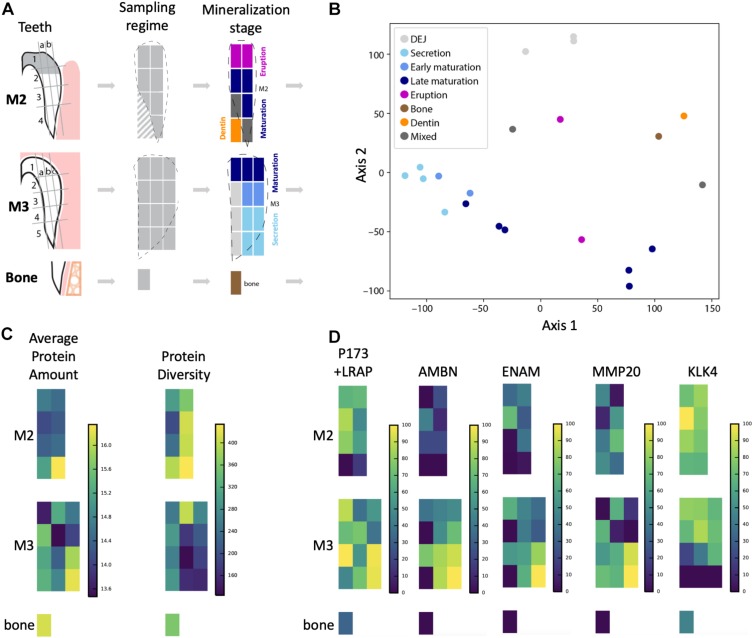
FIGURE 1.
Overview of sampling strategy and stages of enamel mineralization in the forming tooth crown. (A) Porcine molars that were partially erupted (M2) or unerupted and still in the process of crown extension (M3) were dissected and enamel diced into sample blocks, dentin and bone sampled for comparison, and proteins extracted for LC-MS/MS analysis. Dotted lines show outlines of enamel edges relative to diced sample blocks, which are color-coded according to a priori anatomical estimates of tissue and mineralization stage. Diagonal hashes show where dentin tissue was sampled. (B) Principal components analysis shows that samples from secretory enamel sample locations cluster tightly together (light blue), while early and late maturation (darker blue) cluster more loosely. Bone (brown) and dentin (orange) locations cluster separately, as do DEJ samples (light gray). (C) Average peptide amount (peak area) is highest in secretory enamel, while peptide diversity is higher in maturation and erupted enamel. High protein abundance is shown by bright yellow coloration and low abundance by dark blue on the left. High protein diversity is shown by bright yellow and low diversity by dark blue on the right. (D) Abundance for the five traditional amelogenesis proteins, measured as the natural log of the peptide LC-MS/MS peak area and shown on a percent (0–100%) scale per protein, where bright yellow corresponds to high, and dark blue corresponds to low abundance. As expected based on previous observations for these proteins, we observe that AMELX, AMBN, ENAM, and MMP20 have highest abundances in secretory enamel, whereas we do not observe KLK4 in secretory enamel and instead find it in maturing and mature enamel. The spatial pattern seen for the amelogenin P173+LRAP is more complex because we simultaneously track all amelogenin P173+LRAP derived proteins exhibiting the N-terminal sequence (e.g., P173, P56, and their N-terminal derived processing products including P162, P148, P62/3, P45 or TRAP, LRAP, and P40).