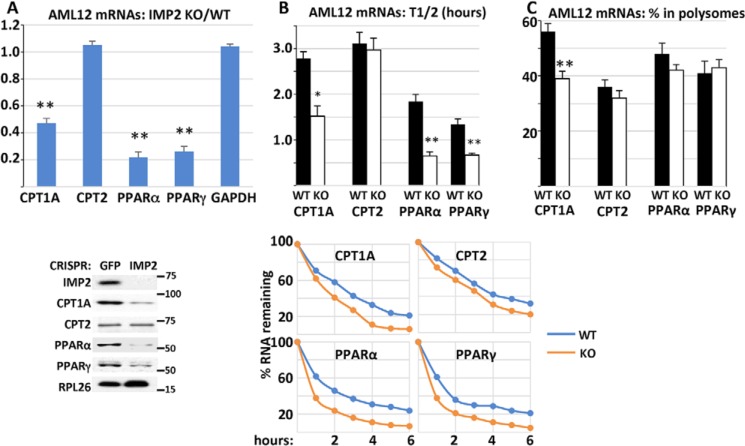
Figure 3.
The effect of Imp2 inactivation in AML12 hepatocytes on the overall abundance, half-life, and polysomal abundance of selected RNAs. A, the effects of CRISPR-catalyzed Imp2 inactivation in AML12 hepatocytes on the abundance of selected mRNAs and polypeptides. RNA was extracted from AML12 hepatocytes transduced with CRISPR reagents directed at Imp2 or GFP, and the abundance of the indicated mRNAs was estimated by QPCR. The ratio of IMP2 CRISPR/GFP CRISPR is shown. Bottom panel, immunoblots of IMP2 and other polypeptides. B, mRNA half-life in IMP2 CRISPR and GFP CRISPR AML12 cells. Cells were treated with actinomycin (1 μm) for 12 h. RNA was extracted at t = 0 and every 2 h thereafter, and the abundance of the indicated mRNAs was determined by QPCR. The rate of decline was plotted by least squares using the first three time points shown in the bottom panel, and the time to 50% decrease of the initial value is shown in the top panel. C, the percentage of mRNA residing in polysomes in IMP2 CRISPR/GFP CRISPR AML12 cells. Total RNA and post-mitochondrial extracts were prepared from equal numbers of rapidly growing cells. The post-mitochondrial extracts were subjected to density gradient centrifugation. Total RNA and RNA from the pooled polysomal fraction of the gradients were quantified by QPCR, and the ratio of polysomal RNA/total RNA × 100 is shown. *, p < 0.05; **, p < 0.01.