Abstract
Diabetes is characterized by a loss of β-cell mass, and a greater understanding of the transcriptional mechanisms governing β-cell function is required for future therapies. Previously, we reported that a complex of the Islet-1 (Isl1) transcription factor and the co-regulator single-stranded DNA–binding protein 3 (SSBP3) regulates the genes necessary for β-cell function, but few proteins are known to interact with this complex in β-cells. To identify additional components, here we performed SSBP3 reverse–cross-linked immunoprecipitation (ReCLIP)- and MS-based experiments with mouse β-cell extracts and compared the results with those from our previous Isl1 ReCLIP study. Our analysis identified the E3 ubiquitin ligases ring finger protein 20 (RNF20) and RNF40, factors that in nonpancreatic cells regulate transcription through imparting monoubiquitin marks on histone H2B (H2Bub1), a precursor to histone H3 lysine 4 trimethylation (H3K4me3). We hypothesized that RNF20 and RNF40 regulate similar genes as those regulated by Isl1 and SSBP3 and are important for β-cell function. We observed that Rnf20 and Rnf40 depletion reduces β-cell H2Bub1 marks and uncovered several target genes, including glucose transporter 2 (Glut2), MAF BZIP transcription factor A (MafA), and uncoupling protein 2 (Ucp2). Strikingly, we also observed that Isl1 and SSBP3 depletion reduces H2Bub1 and H3K4me3 marks, suggesting that they have epigenetic roles. We noted that the RNF complex is required for glucose-stimulated insulin secretion and normal mitochondrial reactive oxygen species levels. These findings indicate that RNF20 and RNF40 regulate β-cell gene expression and insulin secretion and establish a link between Isl1 complexes and global cellular epigenetics.
Keywords: diabetes, transcription factor, transcription regulation, histone modification, chromatin immunoprecipitation (ChiP), insulin secretion, pancreas, islet, H2Bub1, H3K4me3
Introduction
Pancreatic β-cells are at the epicenter of type 1 (cell death) and type 2 (cell dysfunction, de-differentiation, and death) diabetes pathophysiology. Numerous studies have demonstrated that transcription factors (TFs)3 are necessary for proper pancreatic β-cell development and function (1–7). Studies from our group and others have highlighted the importance of the LIM-homeodomain (HD) TF Islet-1 (Isl1), which is expressed in the embryonic pancreatic epithelium and surrounding mesenchyme and is then restricted to adult islets (8–10). Conditional Isl1 removal specifically in late mouse islet development (>embryonic day 13.5) revealed Isl1 roles in maturation of nascent pancreatic endocrine cells. These mice develop postnatal diabetes, attributed to a loss of hormone-positive α-, β-, and δ-cells (11). Tamoxifen-inducible models of adult β-cell Isl1 loss demonstrated that Isl1 is also required for maintaining mature β-cell function. Adult mice lacking Isl1 had impaired glucose tolerance and glucose-stimulated insulin secretion, due in part to dysregulation of Isl1 β-cell targets Pdx1 and Glut2 (12, 13). Transcriptome and lineage-tracing analyses revealed numerous down-regulated transcripts associated with β-cell function, including Ins1, Ins2, Slc2a2, Glp1R, MafA, and Pdx1. Interestingly, Ngn3, an endocrine progenitor TF, was up-regulated in Isl1-depleted β-cells. Although these studies highlight the crucial role of Isl1 in the developing and adult β-cell, the transcriptional mechanisms by which Isl1 complexes impart gene regulation remain unknown.
Despite the powerful impacts of TFs on gene expression, cell differentiation, and function, TFs often require interacting co-regulators to exert influence over transcriptional activation or repression (14, 15). Co-regulators can modulate chromatin structure through post-translational modifications of histones and also direct modification of DNA (e.g. methylation), or simply act as transcriptional scaffolds. For example, the master regulator of pancreas development and β-cell identity and function, Pdx1 (3, 16–18), interacts with the Set7/9 methyltransferase co-regulator to mediate histone modifications and expression of Pdx1 target genes, including Glut2 and insulin I/II (19). Pdx1 also recruits components of the Swi/Snf ATP-dependent chromatin-modifying complex in a glucose-dependent manner to govern β-cell gene expression (20). We hypothesized that, like Pdx1, Isl1 interacts in larger transcriptional complexes to regulate target genes. Indeed, we reported that Isl1 interacts with the scaffolding co-regulator, LIM domain–binding protein 1 (Ldb1), in β-cells (21). Comparison of Isl1 and Ldb1 loss–of–function models revealed similar embryonic and adult β-cell phenotypes, including dysregulation of key TF genes MafA and Arx (13, 21), supporting that these factors cooperatively function to regulate β-cell development and function. Recently, we discovered that the ssDNA-binding protein 3 (SSBP3) co-regulator interacts with Isl1 and Ldb1 and is important for β-cell function (22). siRNA-mediated knockdown and ChIP studies revealed that SSBP3 regulates and occupies the Isl1/Ldb1 target genes, MafA and Glp1R. In this study, our goal was to employ an unbiased reversible cross-link immunoprecipitation (ReCLIP) coupled with mass spectrometry (MS) to compare and characterize the functional protein interactors of the LIM transcriptional complex components Isl1 and SSBP3. We performed an SSBP3 ReCLIP–MS experiment and then compared the dataset with our previously reported Isl1 results (22). We found the ring finger ubiquitin ligase complex components, Rnf20 and Rnf40, in the MS datasets. These factors (homologous to yeast Bre1) are required to modify histone H2B with a monoubiquitination mark (H2Bub1) at lysine 120 (Lys-120) to impact the expression of highly-expressed genes (23, 24), which is also a prerequisite for additional histone modifications, including H3K4 trimethylation (H3K4me3) and H3K79 methylation (H3K79me). Additional analyses found that the expression of β-cell genes, including Glut2, MafA, and Ucp2, were impacted by Rnf complex depletion in vitro, leading to reduced glucose-stimulated insulin secretion (GSIS). Overall, this study sheds light on mechanisms underlying how Isl1 regulates transcription and revealed for the first time that the Rnf20/Rnf40 epigenetic complex impacts β-cell gene expression and function.
Results
ReCLIP reveals β-cell interactions between Rnf20/Rnf40 and the LIM domain complex components Isl1 and SSBP3
To better understand the underlying mechanisms governing the transcriptional control exerted by Isl1, we set out to compare the “interactome” of Isl1 to that of a closely-associated co-regulator (22), ssDNA-binding protein 3 (SSBP3). To accomplish this, we performed an SSBP3 ReCLIP using nuclear extracts from the βTC3 β-cell line, and we compared the resulting MS dataset with that of a species-matched IgG (as negative control) and our previously-reported Isl1 ReCLIP (22). A direct comparison of SSBP3 and Isl1 datasets revealed that 23 proteins were enriched by SSBP3 and Isl1; 121 were SSBP3-specific, and 242 were Isl1-specific (Fig. 1A and Table S1). Among the factors found in the SSBP3 and/or Isl1 datasets were the ring finger ubiquitin ligases Rnf20 and the closely-related Rnf40 (Table S1). These factors form a heterodimeric complex responsible for histone H2B monoubiquitination (H2Bub1), an epigenetic mark associated with gene expression in multiple cell types (23, 24). Interestingly, LIM complex components Isl1, SSBP3, and Ldb1 co-migrated with Rnf20 and Rnf40 in a sucrose gradient into fractions often containing very large proteins and/or complexes (Fig. 1B) (22). To confirm endogenous protein interactions with Rnf20 and Rnf40, we performed co-immunoprecipitations (co-IP) using independently-prepared noncross-linked βTC3 and human islet nuclear extracts. In the Isl1 IP samples, we specifically recovered Rnf20 and Rnf40 (Fig. 1C); however, we did not observe Rnf20 or Rnf40 enrichment in SSBP3 co-IP eluates (data not shown), which may suggest a technical issue with antibody epitope availability or that DSP cross-linking allowed for interaction in ReCLIP samples, but in the absence of cross-links, the binding with SSBP3 is labile or indirect. Nonetheless, we confirmed endogenous Isl1–Rnf20/40 interactions in mouse β-cell lines and in primary human islet extracts (Fig. 1C).
Figure 1.
Ring finger ubiquitin ligases are expressed throughout the developing and adult islet and interact with Isl1. A, Venn diagram comparing independent Isl1 and SSBP3 ReCLIP/MS–enriched proteins (over IgG control). Notably, Rnf20/40 were among the 23 factors common to both datasets. B, sucrose gradient fractionation followed by western blotting of the isolated protein fractions reveals that Isl1, SSBP3, and the Ldb1 co-regulator co-migrate with Rnf20 and Rnf40 factors in high molecular weight fractions. C, confirmatory co-IPs were performed in the absence of cross-linking. Endogenous Isl1 was pulled down, and eluates were blotted with Rnf20- or Rnf40-specific antibodies from βTC3 (left) or human islet (right) nuclear extracts, as compared with positive input control and IgG co-IP as negative control (n = 3). D, western blotting for Rnf20 and Rnf40 in β-cell lines βTC3, Min6, and Ins-1, as compared with NIH3T3 mouse fibroblast cells. Actin is shown as loading control. E, confocal immunofluorescence imaging for Rnf40 (red), Isl1 (blue), and insulin (green) in WT E15.5 pancreas. F, postnatal day (P)1 immunofluorescence demonstrates pancreatic Rnf40 (red) co-staining with Isl1 (blue) and insulin (green). G, section of 3-month-old (3 m) mouse pancreas demonstrating co-expression of Rnf40 (red) and Isl1 (blue) and insulin (green). H, immunofluorescence image of 5-month-old (5M) mouse pancreas section demonstrating broad expression of the histone H2B monoubiquitination mark (H2Bub1, red) throughout the pancreatic field, especially in insulin+ cells (green). I, immunofluorescence image of agarose-embedded nondiabetic human donor islet stained for insulin (green), H2Bub1 (red), and DAPI (blue) to mark nuclei. White arrows point to regions where there were insulin/H2Bub1 co-positive cells. Scale bars (white), 50 μm. Approximate kDa protein sizes are shown to the right of each blot image in B–D.
To understand the relative spatio-temporal expression of the Rnf factors in pancreas tissue and β-cell lines, we employed western blotting and tissue immunofluorescence. We assessed relative Rnf20 and Rnf40 levels in various cell line extracts, and as expected, the Rnf factors are abundantly expressed across all cell lines examined and most notably for this study in βTC3, Min6, and Ins1 β-cell lines (Fig. 1D). In developing embryonic day 15.5 pancreas tissue, Rnf40 was co-expressed in Isl1+ and insulin+ endocrine cells as well as in Ngn3+ (and Isl1− (25)) endocrine progenitors (Fig. 1E and Fig. S1A). This co-staining of Rnf40 with Isl1 and insulin was maintained in postnatal day (P)1 and in 3-month-old adult mouse islets (Fig. 1, F and G). We were unable to perform Rnf20 immunostaining because of a lack of suitable antibody, and SSBP3–Rnf40 co-staining was not possible because the antibodies were raised in the same species (rabbit), thus serial sections were used to suggest similar SSBP3 and Rnf40 expression in P1 β-cells (Fig. S1B). Because the histone H2B monoubiquitination mark imparted by the Rnf complex is largely uncharacterized in islets, we examined the expression pattern by immunofluorescence. As hypothesized, we observed broad H2Bub1 signals throughout the entire pancreatic field, most notably in adult mouse islet cells (Fig. 1H), and in human donor islet β-cells (Fig. 1I).
Loss of Rnf20 and/or Rnf40 impacts expression of key β-cell genes
To begin understanding the functional importance of the Rnf complex in β-cells, we employed an siRNA knockdown approach. Min6 β-cells were transfected with siRNAs directed to Rnf20 or Rnf40 (or a scrambled control), and by western blotting we found global reductions in histone H2B monoubiquitination (H2Bub1) levels, as compared with nontargeting scrambled siRNA (Fig. 2A). These data are in accord with the literature's description of Rnf complex roles in other cell types. Interestingly, we also observed an inter-dependence of Rnf proteins, as reduction of Rnf20 appeared to impact protein levels of Rnf40, and vice versa (Fig. 2A), suggesting a mutual regulation of the ring finger complex components at the protein level, as reported previously (26).
Figure 2.
Rnf20 and Rnf40 are required for monoubiquitination of histone H2B and the expression of β-cell genes. A, western blotting of nuclear extracts from siRNA-transfected (control siSCR, siRnf20, and siRnf40) Min6 cells. Results demonstrate an overt reduction of H2Bub1 in Rnf knockdown extracts, as compared with actin-loading control. B, relative mRNA quantification of various transcriptional effector genes in Min6 cells transfected with control siSCR (black bars, set to 1-fold), siRnf20 (gray bars), or siRnf40 (white bars). C, relative mRNA quantification of various insulin secretion factors in Min6 cells transfected with control siSCR (black bars, set to 1-fold), siRnf20 (gray bars), or siRnf40 (white bars). D, western blotting of MafA and actin loading control from Min6 cells transfected with siSCR, siRnf20, or siRnf40. Interestingly, MafA was marginally elevated upon Rnf20 loss but greatly reduced in the Rnf40 knockdown cell extracts, as demonstrated by densitometry plots. E, western blotting of Ucp2 and actin loading control from Min6 cells transfected with siSCR, siRnf20, or siRnf40. Densitometry quantification revealed significant reduction of Ucp2 protein in the Rnf knockdown whole-cell extracts. F, Glut2 western blotting from Min6 cells transfected with siSCR, siRnf20, or siRnf40, as compared with actin loading control. Densitometry highlights a significant reduction of Glut2 protein in the Rnf knockdown whole-cell extracts. G, relative mRNA quantification of various insulin secretion factors in dispersed primary mouse islet cells transfected with control siSCR (black bars, set to 1-fold), siRnf20 (gray bars), or siRnf40 (white bars). *, p < 0.05; **, p < 0.01; ***, p < 0.001; ****, p < 0.0001. n = 3–5. Approximate kDa protein sizes are shown to the right of each blot image in A and D–F.
To reveal which β-cell genes are dependent on Rnf20 and/or Rnf40 for expression, we performed the Min6 siRNA-mediated knockdown and then assessed β-cell candidate mRNAs by qPCR. In addition to demonstrating the specificity of the siRNAs against Rnf20 or Rnf40, we found that the MafA maturation TF mRNA and protein were significantly reduced upon Rnf40 depletion (Fig. 2, B and D). Interestingly, we did observe a slight but significant elevation of MafA protein upon Rnf20 depletion. This differential effect on MafA suggested to us that the Rnf complex components may have unique function alone, although in light of the observed interdependency of Rnf protein levels, this Rnf-specific impact deserves further attention, especially in vivo. Notably, we observed no change in Pdx1, Isl1, or Ldb1 mRNA levels. Furthermore, glucose-stimulated insulin secretion factors Ins1, Glut2, and the mild uncoupler Ucp2-encoding mRNA were significantly reduced by both Rnf siRNA treatments (Fig. 2C). We also observed a mild reduction of Ucp2 protein in siRnf20- or siRnf40-treated cells (Fig. 2E), suggesting that the Rnf complex regulates Ucp2 to impact β-cell metabolism. We confirmed a significant reduction of Glut2 protein upon Rnf20 or Rnf40 knockdown in Min6 cells (Fig. 2F). The mRNA encoding Glp1R, the insulinotropic incretin receptor on the β-cell, was differentially regulated by Rnf20 and Rnf40, with Rnf20 deficiency imparting a reduction of Glp1r, and Rnf40 leading to an increase. Interestingly, MafA and Glut2 are also known targets of Isl1 and SSBP3 (11, 12, 21, 22), suggesting cooperative control of these genes by the Rnf factors and the LIM-domain transcriptional complex. As a step toward investigating the Rnf20 and Rnf40 importance in primary islet cells, we employed dispersed WT mouse islets for siRNA-mediated knockdown of Rnf20 or Rnf40. Indeed, we observed many similar effects as first observed in Min6 cells, namely reductions of Ins1 and Glut2 levels (Fig. 2G).
Rnf20 and Rnf40 occupy regulatory domains at MafA and Glut2 loci
Next, we wanted to examine whether the Glut2 locus is directly bound by Rnf factors, given that this gene was impacted by loss of Rnf20 and Rnf40 (Fig. 2C). Thus, ChIP using βTC3 β-cell line chromatin was performed. We were able to obtain a ChIP-suitable Rnf20 antibody, but not Rnf40. Because the Rnf complex is known to enrich for proximal or nearby promoters (as opposed to distal promoters/enhancers (27, 28)), we focused our efforts on the upstream promoter domain of Glut2, ∼500–700 bp upstream (Fig. 3A) (29), and the MafA region 6 conserved domain (Fig. 3D, top), which encompasses the transcriptional start site (TSS) (30). Indeed, we observed Rnf20 occupation of MafA region 6 (Fig. 3D, bottom) and the Glut2 promoter, as compared with IgG ChIP control (Fig. 3, A and B). Furthermore, a scanning ChIP (31) across Glut2-coding sequences also revealed H2Bub1 enrichment of exons 1, 4, and 10 (approximating the beginning, middle, and end of the Glut2 locus), with a peak occupancy in the middle of the gene (i.e. exon 4, Fig. 3C), as also noted in the literature for the p21 gene in HCT116 human colorectal carcinoma cells (32). Overall, these data support that Rnf-sensitive β-cell genes, like MafA and Glut2, are occupied by the Rnf complex and that at least the multiexon Glut2 gene is enriched by H2B monoubiquitination in a pattern similar to other highly-expressed genes.
Figure 3.
Rnf20 and the resulting H2Bub1 modification enrich at β-cell gene loci. A, schematic of the mouse Slc2a2/Glut2 locus. Of note is the upstream proximal promoter domain (Prox, gray box), and exonic sequences within the gene body (black boxes), especially the beginning (Exon 1), middle (Exon 4), and end of the gene (Exon 10). B, relative ChIP enrichment by IgG (black bar, set as 1-fold) or Rnf20 (gray bar) of the Glut2 proximal promoter domain. C, relative ChIP enrichment of the H2Bub1 modification throughout the Glut2 gene body. The enrichment values are normalized to IgG control (set to 1-fold). Of note, the enrichment pattern fits a “bell-shaped curve” for H2Bub1, as has been shown in other studies at the p21 locus (32). D, ChIP enrichment of Rnf20 (gray) at the region 6 domain of MafA, a sequence encompassing the proximal promoter (30), as compared with IgG (black), set at 1-fold. *, p < 0.05; ***, p < 0.001; ****, p < 0.0001. n = 3–5.
Isl1 and SSBP3 are required for histone H2B monoubiquitination and histone H3 lysine 4 tri-methylation marks
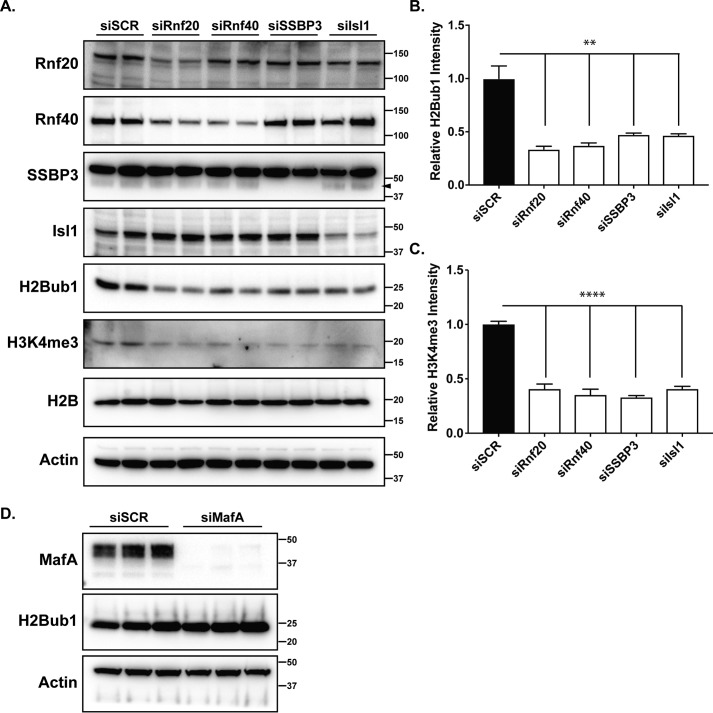
To establish a mechanistic link between the Isl1/SSBP3 complex and the Rnf factors, we set out to define the impacts of Isl1 and SSBP3 on histone ubiquitination and methylation marks. To do this, we returned to the Min6 cells transfected with various siRNAs. As expected, cells transfected with siRNAs against Rnf20 or Rnf40 had reduced H2Bub1 and H3K4me3 marks, as assessed by western blotting (Fig. 4A–C). Strikingly, Isl1- and SSBP3-deficient cells also had reduced steady-state levels of H2Bub1 and H3K4me3 (Fig. 4, A–C), whereas total histone H2B (and actin) levels were unchanged. This effect on H2Bub1 by Isl1 and SSBP3 appears to be specific, as a similar Min6 knockdown of MafA did not impart a reduction of H2Bub1 levels (Fig. 4D). To confirm our observations, we repeated siRNA transfection and western blotting experiments in the rat Ins-1 β-cell line, and observed similar results for H2Bub1 impacts by Isl1 and SSBP3 (Fig. S2).
Figure 4.
Isl1 and SSBP3 are required for H2Bub1 and downstream H3K4me3 modifications in β-cells. A, western blotting of various transcriptional effectors or histone modifications in Min6 cells treated with control siSCR or siRNAs targeting Rnf20, Rnf40, SSBP3, or Isl1. Of note, there was no observed change in total H2B but an overt decrease in H2Bub1 and H3K4me3 modifications. These experiments were performed in duplicate on at least three different occasions. The arrowhead points to the faster-migrating SSBP3-specific band, as opposed to the robust nonspecific band, which we also observed in a prior study (22). Densitometry quantification of H2Bub1 (B) or H3K4me3 (C) intensity across the various siRNA treatments. D, western blotting of siSCR- or siMafA-transfected Min6 cell extracts demonstrating protein levels of MafA (top) and H2Bub1 (middle), as compared with actin loading control (bottom). There was no loss of H2Bub1 levels upon MafA reduction. **, p < 0.01; ****, p < 0.0001. n = 3–4. Approximate kDa protein sizes are shown to the right of each blot image in A and D.
Rnf factors are required to mitigate mitochondrial ROS
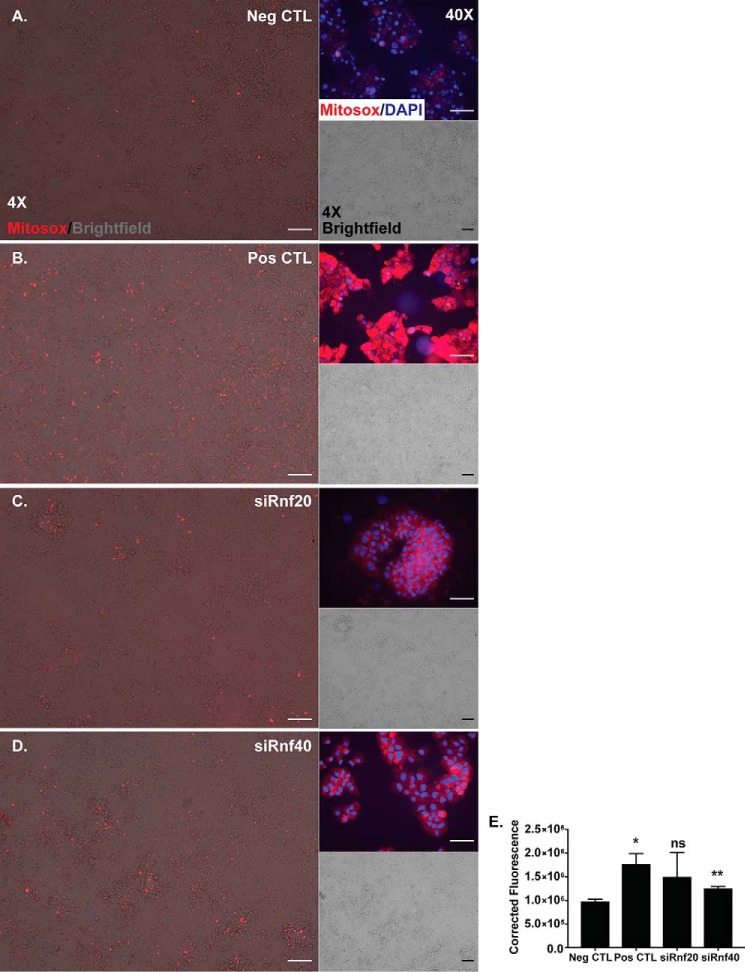
Because of the decrease in Ucp2 mRNA and protein observed upon Rnf knockdowns (Fig. 2, C and E), we next tested for alterations in cellular metabolism. Ucp2 is a mild uncoupler that controls levels of mitochondrial ROS (33–35); thus, we hypothesized that upon Rnf depletion, the reduction of Ucp2 will impart an increase in Min6 ROS levels. Indeed, as compared with Min6 control knockdown cells (siSCR and Neg CTL) and antimycin-treated cells to induce ROS production (Pos CTL), Rnf40 knockdown imparted a significant increase in fluorescent intensity of a mitochondrial-specific ROS dye, MitoSox (Fig. 5), suggesting that at least Rnf40 is required for controlling β-cell ROS levels, a known effector of insulin secretion (36).
Figure 5.
Rnf40-depleted Min6 cells have increased mitochondrial reactive oxygen species. Mitosox staining of Min6 cells to assess mitochondrial ROS is shown. A, siSCR-transfected cells serving as a negative control (Neg CTL) for ROS production. B, antimycin A-treated cells were included as a positive control (Pos CTL) for induction of mitochondrial ROS. siRnf20-transfected (C) or siRnf40-transfected Min6 cell imaging (D) is shown. For each panel, a 4× Mitosox/bright field overlay was included with a corresponding 4× bright field–only panel, plus an independent 40× image of the Mitosox signal merged with DAPI nuclear signal. E, quantification of corrected fluorescence (with ImageJ from at least three independent 4× cell fields) for each treatment group reveals that there is significantly more Mitosox fluorescence in the positive controls and in cells deficient for Rnf40; n = 3. *, p < 0.05; **, p < 0.01; ns, not significant. Scale bars in 4× images (white, black) = 100 μm; Scale bars (white) in 40× images = 50 μm.
Rnf20 and Rnf40 are required for insulin secretion in vitro
As we observed reductions of notable insulin secretion effector genes MafA, Glut2, and Ucp2 (Fig. 2), we sought to test for insulin secretory consequences of Rnf20, Rnf40, and SSBP3 factor loss in β-cells, as compared with Isl1. To do this, we transfected Min6 cells with Rnf20, Rnf40, Isl1, or SSBP3 siRNAs, prior to performing static insulin secretion experiments. Forty eight hours post-transfection, we observed a reduction of high-glucose–stimulated (i.e. 16.7 mm) insulin secretion in Rnf20, Rnf40, and SSBP3-deficient cells (Fig. 6). Isl1 is known to impact insulin secretion (12); thus, there was an expected reduction of GSIS in siIsl1 Min6 cells. Overall, these data highlight that the LIM complex factors (comprised of Isl1 and SSBP3) and Rnf factors are required for GSIS, and it links histone ubiquitination to β-cell function.
Figure 6.
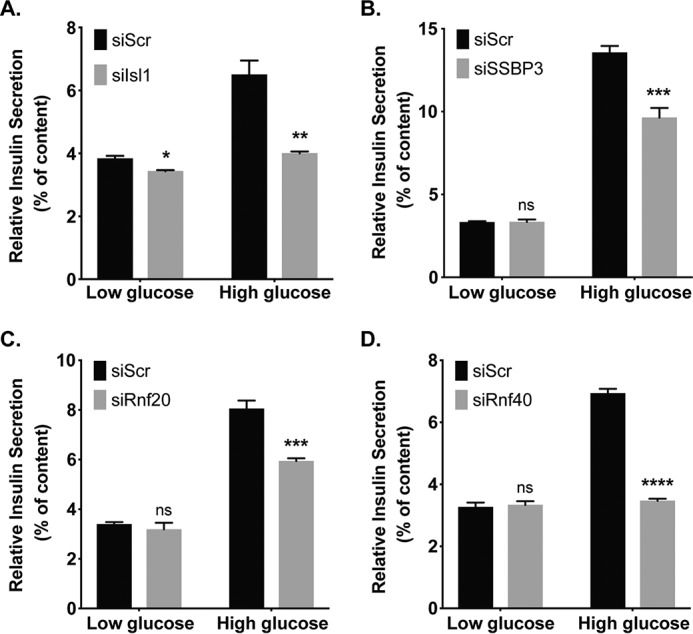
Isl1, SSBP3, and the Rnf factors are necessary for normal glucose-stimulated insulin secretion from β-cells. Static glucose-stimulated secretion assays performed in Min6 cells transfected with a scrambled siRNA (siScr) or siIsl1 (A), siSSBP3 (B), siRnf20 (C), or siRnf40 (D). Results are presented as secreted insulin expressed as percent of insulin content, as measured by ELISA. Data are a mean ± S.E., of duplicate wells repeated on at least three separated occasions. ns, not significant; *, p < 0.05; **, p < 0.01; ***, p < 0.001; ****, p < 0.0001.
Discussion
Dozens of transcription factors are known to be requisite activators and/or repressors involved in pancreatic β-cell development and function (6, 7), and there is growing appreciation for interacting transcriptional co-regulators in the islet field (19, 20, 22, 37–41). Simple TF–DNA interactions (i.e. TF-binding cognate elements) cannot account for all cellular responses to extrinsic and intrinsic stimuli. Hence, additional complexity exists, which is accomplished in part by epigenetic mechanisms employed to elicit precise temporal and cell type-specific gene expression (42, 43). These can occur through various post-translational histone modifications (i.e. methylation and acetylation), noncoding RNAs, and via methylation of CpG islands in genomic DNA (42). Epigenetic mechanisms have become attractive targets for understanding how terminally differentiated β- or α-cells may be coaxed into trans-differentiating toward another islet cell fate. For example, Bramswig et al. (44) found that chemically inhibiting histone methyltransferase levels in human islets, which are required for H3K4me3 (activating) and H3K27me3 (repressive) marks, allowed for an unusual co-expression of insulin and glucagon, as well as Pdx1 and glucagon, suggestive of α–to–β-cell fate conversion.
Although other Isl1 interactors have been described in β-cells, including the previously discussed Ldb1 and SSBP3, the NeuroD1 and Hnf4α TFs (45, 46), and protein inhibitor of activated STAT Y (PIASy) (47), there remains a lack of mechanistic knowledge of how Isl1-mediated complexes impart gene regulatory activity. In this study, we compared endogenous interactions of the Isl1 TF and the SSBP3 co-regulator using β-cell extracts without employing epitope-tagged bait proteins to test the hypothesis that Isl1 and SSBP3 participate in larger transcriptional complex(es) to control gene targets. Of note in the ReCLIP–MS datasets was the presence of the ring finger ubiquitin ligase heterodimeric partners, Rnf20 and Rnf40. Mammalian Rnf20 and Rnf40 act to monoubiquitinate histone H2B (H2Bub1), an activating mark preceding H3K4me3 and H3K79me in cells (48). Thus, the H2Bub1 mark modulates gene expression, rather than histone protein stability, as is the case for polyubiquitin marks. The Rnf complex and H2Bub1 mark have been implicated in numerous cellular processes, including transcriptional initiation and elongation, DNA damage repair, and RNA processing (23). Rnf20 and Rnf40 are generally associated with highly-expressed genes; however, there are examples of target genes being activated or repressed by Rnf factors (49), as we observed for Glp1R upon Rnf40 depletion. Rnf occupation of target loci occurs proximal to TSS (27, 28), whereas H2Bub1 selectively enriches within gene bodies. It is postulated that gene body enrichment of H2Bub1 allows for ejection of histone H2A/H2B histone dimers, allowing pol II to pass through, and may also be required for nucleosome reassembly after pol II passage (50–52). Overall, Rnf20 and Rnf40 are required for monoubiquitination of histone H2B, imparting other histone modifications such as H3K4 trimethylation and H3K79 methylation (53–55) and, ultimately, gene expression.
We confirmed endogenous interaction of Rnf20 and Rnf40 and Isl1 in β-cell lines and human islet extracts, but we were unable to demonstrate interaction between Rnf and SSBP3 in the absence of the DSP cross-linker. This may suggest that the interaction between SSBP3 and Rnf20 observed in the ReCLIP dataset is spurious (Table S1) or that SSBP3 binds only weakly to the Rnf complex, and Isl1 is the major partner. However, we observed novel SSBP3 impacts on H2B monoubiquitination, H3K4 tri-methylation, and GSIS, suggesting that SSBP3 indeed participates in Rnf–Isl1-mediated complexes or at least acts through known effects on LIM-factor stability (e.g. Isl1) (22, 56). We also observed no interaction of the Rnf complex components with the LIM co-regulator, Ldb1 (13, 21), again suggesting that the Rnf factors may be Isl1-specific partners in β-cells (data not shown).
Using immunofluorescence, we found that Rnf40 broadly decorated the developing and adult pancreas, including Ngn3+ endocrine progenitors and insulin+ islet cells. This suggested to us that the Rnf complex could have developmental roles in islets as well, with possible implications for Rnf factors in the directed differentiation of ES cells toward β-like cells in vitro (57). We found that global H2Bub1 marks were reduced in Min6 cells deficient in Rnf20 or Rnf40, which is in accord with roles described in other cell/tissue types (32, 48, 58). Our knockdown analyses revealed that MafA (an Isl1, Ldb1, and SSBP3 target gene (11, 21, 22)), Glut2 (also an Isl1 target gene (12)), and Ucp2 mRNA levels were sensitive to Rnf depletion. We employed Glut2 as a model multiexon gene for our ChIP studies and observed Rnf20 occupation at the immediate upstream domain of Glut2 (29), whereas the H2Bub1 mark was enriched throughout the coding region, as described for other highly-expressed genes (27). Similar occupation of MafA region 6 (encompassing the TSS (30)) by Rnf20 was observed, suggesting that MafA is a direct target of Rnf complexes. However, because MafA is a single-exon gene, we did not pursue H2Bub1 ChIP studies, similar to Glut2. To date, we have not linked Ucp2 expression in β-cells to control by any LIM-domain (i.e. Isl1-containing) complex.
A striking mechanistic link between Isl1 and the Rnf complex was established in our Min6 knockdown model, as H2Bub1 and H3K4me3 levels were reduced upon Isl1 and SSBP3 loss, leaving us to postulate that Isl1 may traffic the Rnf complex to target gene loci to establish H2Bub1 marks. Furthermore, it was surprising to observe this reduction in global H2Bub1 (but not total H2B) and H3K4me3 levels in Isl1- or SSBP3-deficient cells, and not solely at specific target gene loci (e.g. Glut2 in Fig. 3). This suggested to us that Isl1 is important for establishing this epigenetic mark throughout the β-cell genome, and it may be independent of whether any particular gene is a direct Isl1 target. This is supported by the observation that Rnf20 and Rnf40 mRNA levels were not perturbed in the siIsl1 cells. Alternatively, it is possible that Isl1 complexes are also regulating the expression or activity of histone de-ubiquitinating enzymes, for example Ubp8 and Ubp10 (59), which could impart similar changes in H2Bub1.
Our gene expression analyses pointed to the Ucp2 gene, encoding a mild β-cell uncoupling protein, as a target of Rnf complexes, specifically sensitive to loss of Rnf40. Ucp2 is important for mitigating mitochondrial ROS, and thus β-cell stress (34). We found that reducing Rnf40 levels in Min6 cells imparted a decrease in Ucp2 mRNA and protein, while increasing mitochondrial ROS, as observed by Mitosox staining. This led us to conclude that Rnf factors are required to maintain normal β-cell ROS through regulation of Ucp2. This implies that Rnf complex activity is linked to β-cell health and may be involved in cellular stress pathways. Looking toward the future, it is possible that Rnf complexes are dysregulated and modulate gene expression in mouse models of metabolic stress, for example during high-fat diet, in db/db mice, or in early type 1 diabetes (60). Additional studies will examine Rnf20 and Rnf40 impacts on β-cell stress and dysfunction.
We observed decreased GSIS upon Rnf20/Rnf40 or SSBP3 depletion from Min6 cells, as compared with known Isl1 impacts (12, 61). This suggested to us that the collective effects on target genes, including Glut2, MafA, insulin I, and Ucp2 have functional consequences in β-cells. Notably, we did not observe basal secretory machinery mRNAs altered by (at least) Rnf20 depletion (Fig. S3), perhaps suggesting that the Rnf or Isl1 complexes impact another aspect of GSIS, including glucose-sensing, metabolism, cellular depolarization, or calcium influx.
Taken together, our results on the understudied islet Rnf20/Rnf40 complex and resulting H2Bub1 modification have several implications and unanswered questions. First, we uncovered that Isl1 has novel roles in impacting modified histone levels in β-cells in vitro. It is unclear whether Isl1 is an exclusive interactor of the Rnf complex in β-cells, but as Isl1 is broadly distributed throughout the islet, Isl1 and the Rnf complex may also function together in α-cells. Second, although we observed Rnf impacts on β-cell function in vitro (e.g. GSIS), it will be interesting to explore whether the Rnf complex is also required for maintaining β-cell identity in vivo, as in the adult Isl1 and Ldb1 knockout models or in the Bramswig et al. study of histone methylation in human islets (13, 44). Finally, there are obvious implications for this work on the embryonic development of islet cells and on current ES cell-based differentiation. These approaches likely need consideration of ring-finger ubiquitin ligase action and histone H2B modification status. Future directions will include examining the roles of Rnf20/Rnf40 in vivo during pancreas/islet organogenesis and in adult β-cell function, with the hypothesis that the Rnf ubiquitin ligases and the resulting H2Bub1 mark are required to establish and maintain β-cell identity during islet development and in adult mammals. Although it is tempting to dismiss the Rnf factors and H2Bub1 modification as likely having pleiotropic impacts on gene transcription in any Rnf-deficient cell, the literature may point to a different outcome, as there appears to be selectivity in Rnf action on gene targets (58). For example, it may be advantageous to increase levels of highly-expressed genes (e.g. insulin and Glut2) in ES-derived β-cells via selective Rnf agonism. Upcoming mouse model experiments will examine the necessity of Rnf factors in islet organogenesis to begin answering these in vivo questions. In total, we found that Isl1 transcriptional complexes are also composed of Rnf20 and Rnf40 ubiquitin ligases, which impact histone modifications, gene expression, and β-cell function.
Experimental procedures
ReCLIP and mass spectrometry
Cell cross-linking and IP were performed as described (22). Briefly, for cross-linking, mouse βTC3 cells were incubated with 0.5 mm DSP (Thermo Fisher Scientific) in 1× PBS for 30 min at 37 °C. Nuclear extracts were prepared with a modified nuclear extraction buffer (62, 63), excluding DTT, EDTA, and EGTA. Cross-linked nuclear extracts were incubated with α-SSBP3 (ab83815; Abcam) antibody, or species-matched control IgG cross-linked to protein G Dynabeads (Life Technologies, Inc.) with dimethyl pimelimidate (Life Technologies, Inc.). IP was carried out for 3 h at 4 °C before washing with radioimmunoprecipitation assay (RIPA) buffer (50 mm Tris, pH 7.4, 150 mm NaCl, 1% Nonidet P-40, 0.5% deoxycholic acid, 0.1% SDS) supplemented with protease inhibitor cocktail (PIC) (PC198152, Thermo Fisher Scientific), and then eluting with RIPA buffer supplemented with 150 mm DTT. SSBP3- and IgG-enriched elutions were sent to University of Alabama at Birmingham Mass Spectrometry/Proteomics Consortium for MS analysis, as described previously (22).
Mass spectrometry
SSBP3- and Isl-enriched (and IgG controls) protein eluates were diluted in LDS-PAGE buffer (Invitrogen) followed by reducing, heat denaturation, and separation by 4–12% SDS-PAGE. The gel was stained overnight with colloidal blue (Invitrogen). The entire lane was cut into six molecular weight fractions, and each fraction was analyzed separately by first cutting into small pieces and equilibrated in 100 mm ammonium bicarbonate. The gel pieces were then reduced, carbidomethylated, dehydrated, and digested with Trypsin Gold (Promega) as per the manufacturer's instructions. After digestion, peptides were extracted, concentrated, and resolubilized in 0.1% formic acid before analysis. Peptide digests (8 μl each) were injected onto a 1260 Infinity nHPLC stack (Agilent Technologies) and separated using a 75 μm inner diameter × 15-cm pulled tip C-18 column (Jupiter C-18 300 Å, 5 μm, Phenomenex). This system runs in-line with a Thermo Orbitrap Velos Pro hybrid mass spectrometer, equipped with a nano-electrospray source (Thermo Fisher Scientific), and all data were collected in collision-induced dissociation mode. The nHPLC was configured with binary mobile phases that included solvent A (0.1% FA in ddH2O), and solvent B (0.1% FA in 15% ddH2O, 85% acetonitrile) programmed as follows: 10 min at 5% B (2 μl/min, load), 90 min at 5–40% B (linear, 0.5 nl/min, analyze), 5 min at 70% B (2 μl/min, wash), and 10 min at 0% B (2 μl/min, equilibrate) (64). Following each parent ion scan (300–1200 m/z at 60,000 resolution), fragmentation data (MS2) were collected on the top most intense 15 ions. For data-dependent scans, charge state screening and dynamic exclusion were enabled with a repeat count of 2, repeat duration of 30 s, and exclusion duration of 90 s.
MS data conversion and searches
The XCalibur RAW files were collected in profile mode, centroided, and converted to MzXML using ReAdW version 3.5.1. The mgf files were then created using MzXML2Search (included in TPP version 3.5) for all scans. The data were searched using SEQUEST, which was set for two maximum missed cleavages, a precursor mass window of 20 ppm, trypsin digestion, variable modification C at 57.0293 and M at 15.9949. Searches were performed with a species-specific subset of the UniRef100 database (updated annually).
Peptide filtering, grouping, and quantification
The list of peptide IDs generated based on SEQUEST (Thermo Fisher Scientific) search results were filtered using Scaffold (Protein Sciences, Portland, OR). Scaffold filters and groups all peptides to generate and retain only high-confidence IDs while also generating normalized spectral counts across all samples for the purpose of relative quantification. The filter cutoff values were set with minimum peptide length of >5 amino acids, with no MH + 1 charge states, with peptide probabilities of >80% C.I., and with the number of peptides per protein ≥2. The protein probabilities were then set to a >99.0% C.I., and a false discovery rate (FDR) of <1.0. Scaffold incorporates the two most common methods for statistical validation of large proteome datasets, the FDR, and protein probability (65–67). Relative quantification across experiments were then performed via spectral counting (68, 69), and when relevant, spectral count abundances were then normalized between samples (70).
Sucrose gradient fractionation
Sucrose gradients were prepared as described previously (22, 38). βTC3 nuclear extracts were prepared and fractionated overnight at 50,000 rpm in a TLS-55 swinging bucket rotor at 4 °C through a 10–35% sucrose gradient diluted in DNA-binding buffer (10 mm HEPES, pH 8.0, 100 mm NaCl, 1 mm EDTA, and 2 mm DTT). Eighteen fractions (∼200 μl each) were collected and separated by 10% SDS-PAGE and analyzed by western blotting using the following antibodies: α-Ldb1 (1:1000, sc-11198; Santa Cruz Biotechnology); α-Isl1 (1:1000, 39.4D5, Developmental Studies Hybridoma Bank (DSHB)); α-SSBP3 (1:500, ab83815; Abcam); α-Rnf20 (1:1000, A300-714a Bethyl Laboratories); α-Rnf40 (1:1000, ab191309, Abcam).
Co-immunoprecipitation and western blotting
α-Isl1 (39.4D5, DSHB) or control IgG antibodies were bound to protein G Dynabeads and then incubated with βTC3 or human islet extracts diluted in PBS supplemented with PIC. Nondiabetic human pancreatic islets were provided by the NIDDK-funded Integrated Islet Distribution Program. Antibody-bound beads were washed with PBS/PIC and eluted with RIPA buffer at 37 °C. Elutions were separated by 10% SDS-PAGE (Bio-Rad) then transferred to a polyvinylidene difluoride (PVDF) membrane. For western blotting (WB), PVDF membranes were blocked in PBS/Tween plus 5% nonfat dry milk for 1 h, followed by an incubation with α-Rnf20 (1:1000, A300-714a, Bethyl Laboratories), α-Rnf40 (1:1000, ab191309, Abcam), α-Isl1 (1:1000, 39.4D5, DSHB), α-SSBP3 (1:1000, ab83815, Abcam), α-MafA (1:1000, NBP1-00121, Novus), α-Ucp2 (1:1000, 89326, Cell Signaling Technology), α-Glut2 (Santa Cruz Biotechnology, sc-7580) antibody overnight at 4 °C. The membrane was washed and incubated with species-matched horseradish peroxidase-conjugated secondary antibodies (Promega or Santa Cruz Biotechnology) followed by addition of Luminata Forte substrate (Millipore) and visualized using ChemiDoc XRS + Imager (Bio-Rad). All co-IP and WB experiments were performed at least three times.
Tissue collection and immunofluorescence
Embryonic and adult mouse pancreata were fixed in 4% paraformaldehyde in 1× PBS for 4 and 8 h, respectively, and then paraffin-embedded. Sections were cut to 6 μm, rehydrated, antigen-retrieved in 1× TEG buffer, and blocked with 5% normal donkey serum in 1% BSA/1× PBS. Sections were incubated in primary antibody overnight at 4 °C: rabbit α-SSBP3 (1:500, ab83815; Abcam); mouse α-Isl1 (1:1000, 39.4D5; DSHB); rabbit α-Rnf40 (1:1000, ab191309; Abcam); mouse α-neurogenin-3 (Ngn3) (1:1000, F25A1B3; DSHB); rabbit α-ubiquityl-histone H2B (1:1000, 5546S, Cell Signaling Technology); guinea pig α-insulin (1:1000, 0564; Dako). Cy-2-, Cy-3-, and Cy-5–conjugated donkey α-rabbit, α-mouse, and α-guinea pig IgG secondary antibodies (1:500; Jackson ImmunoResearch) were used for detection. Slides were imaged using a Zeiss LSM710 confocal microscope or Olympus IX81 fluorescence microscope, and the images were processed by Zen software (Zeiss) or CellSens Dimensions version 1.12 (Olympus) software.
Transient transfection
For small interfering RNA (siRNA) transfection experiments, mouse Min6, rat Ins1 cells, and dispersed mouse islet cells (22, 71) were seeded in 6- or 12-well plates and then transfected with 50 nm On-Target Plus Smart Pool targeting SSBP3 (siSSBP3, L-042856-01-0005), Isl1 (siIsl1, L-059394-01-0005), MafA (siMafA, L-041353-01-0005), Rnf20 (siRnf20, L-041733-01-0005) or Rnf40 (siRnf40, L-059014-01-0005), or nontargeted scrambled control (siSCR, D-001810-GE Healthcare/Dharmacon) using RNAiMax (13778030; Life Technologies, Inc.). WB, mRNA, or insulin secretion analyses were performed 48 h post-transfection.
qPCR
RNA was isolated from Min6 cells using the RNeasy mini plus kit (74134; Qiagen), and cDNA was made using the iScript cDNA synthesis kit (170-8840; Bio-Rad). qPCRs were performed using iTaq SYBR Green (172–5124; Bio-Rad) in duplicate using a LightCycler 480 II (Roche Applied Science) and analyzed using the 2−ΔΔCT method, with normalization to the gapdh housekeeping gene. Primer sequences can be found in Ref. 22 or will be provided upon request.
Chromatin IP (ChIP)
ChIP was performed as described (21, 22, 31). Briefly, βTC3 cells were seeded at 4 × 106 cells per 10-cm dish and cultured for 72 h. Cells were cross-linked with 1% formaldehyde in Dulbecco's modified Eagle's medium at room temperature. Chromatin was fragmented by sonication (Biorupter XL; Diagenode) and precleared with protein G Dynabeads for 2 h at 4 °C. Precleared chromatin was incubated overnight with α-Rnf20 (P120-101; Bethyl Laboratories), α-H2Bub1 (catalog no. 5546, Cell Signaling Technology), or rabbit IgG (Bethyl Laboratories) antibody. Antibody-bound chromatin was incubated with protein G Dynabeads at 4 °C for 3 h. Beads were washed, and chromatin was eluted, and then cross-links were reversed. qPCR was performed on immunoprecipitated DNA using iTaq SYBR Green (Bio-Rad) and a LightCycler 480 II (Roche Applied Science), as described above. Fold enrichment of target sequences in ChIP DNAs were normalized to inactive albumin control sequences and species-matched IgG enrichment (ΔΔCT). Each ChIP experiment was repeated at least three times using independently prepared chromatin. Primer sequences are from Refs. 22, 29, 30 or can be provided upon request.
GSIS
Forty-eight hours after siRNA knockdown or scramble control, Min6 or Ins1 cells were stimulated in low (2.8 mm) or stimulatory (16.7 mm) glucose in Krebs-Ringer bicarbonate/HEPES buffer (KRBH) for 30 min at 37 °C, similar to that described in Ref. 72. Supernatant media, with secreted insulin, were collected. Cells were then lysed using acid/ethanol to obtain total insulin content. Secreted insulin and total insulin were measured by ELISA, and analysis was represented by secreted insulin as % of insulin content. The knockdown-GSIS experiments were repeated on at least three separate occasions.
Visualization and quantification of mitochondrial ROS
Mitochondrial ROS levels were visualized in siRNA-treated Min6 cells using MitoSox staining according to the manufacturer's instructions (Invitrogen, M36008). Fluorescent Mitosox images were compared with bright field to visualize total cell numbers. To quantify the Mitosox signal, we employed ImageJ (National Institutes of Health). Total fluorescence intensity was quantified in at least three independent cellular fields within two duplicate wells, with background correction. Imaging shown is representative of three independent Min6 knockdowns.
Animal use
All studies were approved by and performed according to the guidelines of the University of Alabama at Birmingham Institutional Animal Care and Use Committee.
Statistical analyses
Data are presented as mean ± S.E. Significance was determined after performing a Student's t test, for which p < 0.05.
Author contributions
A. K. W., M. M. B., H. M. T., and C. S. H. conceptualization; A. K. W., Y. L., and C. S. H. data curation; A. K. W., Y. L., M. M. B., E. T., H. M. T., and C. S. H. formal analysis; A. K. W., Y. L., M. M. B., E. T., and C. S. H. investigation; A. K. W. and C. S. H. visualization; A. K. W., Y. L., M. M. B., E. T., H. M. T., and C. S. H. methodology; A. K. W. and C. S. H. writing-original draft; A. K. W., M. M. B., E. T., H. M. T., and C. S. H. writing-review and editing; H. M. T. and C. S. H. resources; C. S. H. supervision; C. S. H. funding acquisition; C. S. H. project administration.
Supplementary Material
Acknowledgment
We thank Jessica Kepple (University of Alabama at Birmingham) for manuscript critiques. The University of Alabama at Birmingham Comprehensive Cancer Center was recipient of National Institutes of Health Grant P30CA013148 from the NCI.
This work was supported by the National Institutes of Health Grants R01DK111483 (to C. S. H.), F31DK111181 (to M. M. B.), T32GM008111 (to E. T.), and R01DK099550 (to H. M. T.) and American Diabetes Association Grants 1-16-JDF-044 (to C. S. H) and 1-17-MUI-004 (to A. K. W. and C. S. H.). The authors declare that they have no conflicts of interest with the contents of this article. The content is solely the responsibility of the authors and does not necessarily represent the official views of the National Institutes of Health.
This article contains Figs. S1–S3 and Table S1.
- TF
- transcription factor
- Rnf
- Ring finger
- LIM
- Lin 11–Isl1–Mec3
- HD
- homeodomain
- H2Bub1
- histone H2B monoubiquitination
- H3K4me3
- histone H3 lysine 4 trimethylation
- H3K79me
- histone H3 lysine 79 methylation
- ROS
- reactive oxygen species
- GSIS
- glucose-stimulated insulin secretion
- Isl1
- Islet-1
- ReCLIP
- reversible cross-link immunoprecipitation
- co-IP
- co-immunoprecipitation
- WB
- western blotting
- PIC
- protease inhibitor cocktail
- PVDF
- polyvinylidene difluoride
- ssDNA
- single-stranded DNA
- FA
- formic acid
- ddH2O
- double-distilled H2O
- qPCR
- quantitative RT-PCR
- TSS
- transcriptional start site
- pol
- polymerase
- DSP
- dithiobis(succinimidyl propionate)
- P
- postnatal
- IP
- immunoprecipitation
- FDR
- false discovery rate
- DSHB
- Developmental Studies Hybridoma Bank
- DAPI
- 4′,6-diamidino-2-phenylindole
- ES
- embryonic stem cell
- C.I.
- confidence interval
- MS
- mass spectrometry.
References
- 1. Gradwohl G., Dierich A., LeMeur M., and Guillemot F. (2000) Neurogenin3 is required for the development of the four endocrine cell lineages of the pancreas. Proc. Natl. Acad. Sci. U.S.A. 97, 1607–1611 10.1073/pnas.97.4.1607 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Hang Y., Yamamoto T., Benninger R. K., Brissova M., Guo M., Bush W., Piston D. W., Powers A. C., Magnuson M., Thurmond D. C., and Stein R. (2014) The MafA transcription factor becomes essential to islet beta-cells soon after birth. Diabetes 63, 1994–2005 10.2337/db13-1001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Jonsson J., Carlsson L., Edlund T., and Edlund H. (1994) Insulin-promoter-factor 1 is required for pancreas development in mice. Nature 371, 606–609 10.1038/371606a0 [DOI] [PubMed] [Google Scholar]
- 4. Offield M. F., Jetton T. L., Labosky P. A., Ray M., Stein R. W., Magnuson M. A., Hogan B. L., and Wright C. V. (1996) PDX-1 is required for pancreatic outgrowth and differentiation of the rostral duodenum. Development 122, 983–995 [DOI] [PubMed] [Google Scholar]
- 5. Zhang C., Moriguchi T., Kajihara M., Esaki R., Harada A., Shimohata H., Oishi H., Hamada M., Morito N., Hasegawa K., Kudo T., Engel J. D., Yamamoto M., and Takahashi S. (2005) MafA is a key regulator of glucose-stimulated insulin secretion. Mol. Cell. Biol. 25, 4969–4976 10.1128/MCB.25.12.4969-4976.2005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Oliver-Krasinski J. M., and Stoffers D. A. (2008) On the origin of the beta cell. Genes Dev. 22, 1998–2021 10.1101/gad.1670808 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Pan F. C., and Wright C. (2011) Pancreas organogenesis: from bud to plexus to gland. Dev. Dyn. 240, 530–565 10.1002/dvdy.22584 [DOI] [PubMed] [Google Scholar]
- 8. Ahlgren U., Pfaff S. L., Jessell T. M., Edlund T., and Edlund H. (1997) Independent requirement for ISL1 in formation of pancreatic mesenchyme and islet cells. Nature 385, 257–260 10.1038/385257a0 [DOI] [PubMed] [Google Scholar]
- 9. Thor S., Ericson J., Brännström T., and Edlund T. (1991) The homeodomain LIM protein Isl-1 is expressed in subsets of neurons and endocrine cells in the adult rat. Neuron 7, 881–889 10.1016/0896-6273(91)90334-V [DOI] [PubMed] [Google Scholar]
- 10. Zhuang S., Zhang Q., Zhuang T., Evans S. M., Liang X., and Sun Y. (2013) Expression of Isl1 during mouse development. Gene Expr. Patterns 13, 407–412 10.1016/j.gep.2013.07.001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Du A., Hunter C. S., Murray J., Noble D., Cai C. L., Evans S. M., Stein R., and May C. L. (2009) Islet-1 is required for the maturation, proliferation, and survival of the endocrine pancreas. Diabetes 58, 2059–2069 10.2337/db08-0987 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Ediger B. N., Du A., Liu J., Hunter C. S., Walp E. R., Schug J., Kaestner K. H., Stein R., Stoffers D. A., and May C. L. (2014) Islet-1 is essential for pancreatic beta-cell function. Diabetes 63, 4206–4217 10.2337/db14-0096 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Ediger B. N., Lim H. W., Juliana C., Groff D. N., Williams L. T., Dominguez G., Liu J. H., Taylor B. L., Walp E. R., Kameswaran V., Yang J., Liu C., Hunter C. S., Kaestner K. H., Naji A., et al. (2017) LIM domain-binding 1 maintains the terminally differentiated state of pancreatic beta cells. J. Clin. Invest. 127, 215–229 10.1172/JCI88016 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Lonard D. M., and O'Malley B. W. (2012) Nuclear receptor coregulators: modulators of pathology and therapeutic targets. Nat. Rev. Endocrinol. 8, 598–604 10.1038/nrendo.2012.100 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Rosenfeld M. G., Lunyak V. V., and Glass C. K. (2006) Sensors and signals: a coactivator/corepressor/epigenetic code for integrating signal-dependent programs of transcriptional response. Genes Dev. 20, 1405–1428 10.1101/gad.1424806 [DOI] [PubMed] [Google Scholar]
- 16. Gao T., McKenna B., Li C., Reichert M., Nguyen J., Singh T., Yang C., Pannikar A., Doliba N., Zhang T., Stoffers D. A., Edlund H., Matschinsky F., Stein R., and Stanger B. Z. (2014) Pdx1 maintains beta cell identity and function by repressing an α cell program. Cell Metab. 19, 259–271 10.1016/j.cmet.2013.12.002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Yang Y. P., Thorel F., Boyer D. F., Herrera P. L., and Wright C. V. (2011) Context-specific α-to-β-cell reprogramming by forced Pdx1 expression. Genes Dev. 25, 1680–1685 10.1101/gad.16875711 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Stoffers D. A., Zinkin N. T., Stanojevic V., Clarke W. L., and Habener J. F. (1997) Pancreatic agenesis attributable to a single nucleotide deletion in the human IPF1 gene coding sequence. Nat. Genet. 15, 106–110 10.1038/ng0197-106 [DOI] [PubMed] [Google Scholar]
- 19. Deering T. G., Ogihara T., Trace A. P., Maier B., and Mirmira R. G. (2009) Methyltransferase Set7/9 maintains transcription and euchromatin structure at islet-enriched genes. Diabetes 58, 185–193 10.2337/db08-1150 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. McKenna B., Guo M., Reynolds A., Hara M., and Stein R. (2015) Dynamic recruitment of functionally distinct Swi/Snf chromatin remodeling complexes modulates Pdx1 activity in islet beta cells. Cell Rep. 10, 2032–2042 10.1016/j.celrep.2015.02.054 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Hunter C. S., Dixit S., Cohen T., Ediger B., Wilcox C., Ferreira M., Westphal H., Stein R., and May C. L. (2013) Islet α-, β-, and δ-cell development is controlled by the Ldb1 coregulator, acting primarily with the islet-1 transcription factor. Diabetes 62, 875–886 10.2337/db12-0952 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Galloway J. R., Bethea M., Liu Y., Underwood R., Mobley J. A., and Hunter C. S. (2015) SSBP3 interacts with islet-1 and Ldb1 to impact pancreatic beta-cell target genes. Mol. Endocrinol. 29, 1774–1786 10.1210/me.2015-1165 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Fuchs G., and Oren M. (2014) Writing and reading H2B monoubiquitylation. Biochim. Biophys. Acta 1839, 694–701 10.1016/j.bbagrm.2014.01.002 [DOI] [PubMed] [Google Scholar]
- 24. Shiloh Y., Shema E., Moyal L., and Oren M. (2011) RNF20-RNF40: a ubiquitin-driven link between gene expression and the DNA damage response. FEBS Lett. 585, 2795–2802 10.1016/j.febslet.2011.07.034 [DOI] [PubMed] [Google Scholar]
- 25. Schwitzgebel V. M., Scheel D. W., Conners J. R., Kalamaras J., Lee J. E., Anderson D. J., Sussel L., Johnson J. D., and German M. S. (2000) Expression of neurogenin3 reveals an islet cell precursor population in the pancreas. Development 127, 3533–3542 [DOI] [PubMed] [Google Scholar]
- 26. Duan Y., Huo D., Gao J., Wu H., Ye Z., Liu Z., Zhang K., Shan L., Zhou X., Wang Y., Su D., Ding X., Shi L., Wang Y., Shang Y., and Xuan C. (2016) Ubiquitin ligase RNF20/40 facilitates spindle assembly and promotes breast carcinogenesis through stabilizing motor protein Eg5. Nat. Commun. 7, 12648 10.1038/ncomms12648 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Wood A., Schneider J., Dover J., Johnston M., and Shilatifard A. (2003) The Paf1 complex is essential for histone monoubiquitination by the Rad6-Bre1 complex, which signals for histone methylation by COMPASS and Dot1p. J. Biol. Chem. 278, 34739–34742 10.1074/jbc.C300269200 [DOI] [PubMed] [Google Scholar]
- 28. Kim J., Guermah M., McGinty R. K., Lee J. S., Tang Z., Milne T. A., Shilatifard A., Muir T. W., and Roeder R. G. (2009) RAD6-mediated transcription-coupled H2B ubiquitylation directly stimulates H3K4 methylation in human cells. Cell 137, 459–471 10.1016/j.cell.2009.02.027 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Chakrabarti S. K., James J. C., and Mirmira R. G. (2002) Quantitative assessment of gene targeting in vitro and in vivo by the pancreatic transcription factor, Pdx1. Importance of chromatin structure in directing promoter binding. J. Biol. Chem. 277, 13286–13293 10.1074/jbc.M111857200 [DOI] [PubMed] [Google Scholar]
- 30. Raum J. C., Gerrish K., Artner I., Henderson E., Guo M., Sussel L., Schisler J. C., Newgard C. B., and Stein R. (2006) FoxA2, Nkx2.2, and PDX-1 regulate islet beta-cell-specific mafA expression through conserved sequences located between base pairs −8118 and −7750 upstream from the transcription start site. Mol. Cell. Biol. 26, 5735–5743 10.1128/MCB.00249-06 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Hunter C. S., Maestro M. A., Raum J. C., Guo M., Thompson F. H. 3rd, Ferrer J., and Stein R. (2011) Hnf1α (MODY3) regulates beta-cell–enriched MafA transcription factor expression. Mol. Endocrinol. 25, 339–347 10.1210/me.2010-0362 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Minsky N., Shema E., Field Y., Schuster M., Segal E., and Oren M. (2008) Monoubiquitinated H2B is associated with the transcribed region of highly expressed genes in human cells. Nat. Cell Biol. 10, 483–488 10.1038/ncb1712 [DOI] [PubMed] [Google Scholar]
- 33. Nicholls D. G. (2016) The pancreatic beta-cell: a bioenergetic perspective. Physiol. Rev. 96, 1385–1447 10.1152/physrev.00009.2016 [DOI] [PubMed] [Google Scholar]
- 34. Robson-Doucette C. A., Sultan S., Allister E. M., Wikstrom J. D., Koshkin V., Bhattacharjee A., Prentice K. J., Sereda S. B., Shirihai O. S., and Wheeler M. B. (2011) Beta-cell uncoupling protein 2 regulates reactive oxygen species production, which influences both insulin and glucagon secretion. Diabetes 60, 2710–2719 10.2337/db11-0132 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Pi J., Bai Y., Daniel K. W., Liu D., Lyght O., Edelstein D., Brownlee M., Corkey B. E., and Collins S. (2009) Persistent oxidative stress due to absence of uncoupling protein 2 associated with impaired pancreatic beta-cell function. Endocrinology 150, 3040–3048 10.1210/en.2008-1642 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Gerber P. A., and Rutter G. A. (2017) The role of oxidative stress and hypoxia in pancreatic beta-cell dysfunction in diabetes mellitus. Antioxid. Redox Signal. 26, 501–518 10.1089/ars.2016.6755 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Qiu Y., Guo M., Huang S., and Stein R. (2002) Insulin gene transcription is mediated by interactions between the p300 coactivator and PDX-1, BETA2, and E47. Mol. Cell. Biol. 22, 412–420 10.1128/MCB.22.2.412-420.2002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Scoville D. W., Cyphert H. A., Liao L., Xu J., Reynolds A., Guo S., and Stein R. (2015) MLL3 and MLL4 methyltransferases bind to the MAFA and MAFB transcription factors to regulate islet beta-cell function. Diabetes 64, 3772–3783 10.2337/db15-0281 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Thomas M. K., Yao K. M., Tenser M. S., Wong G. G., and Habener J. F. (1999) Bridge-1, a novel PDZ-domain coactivator of E2A-mediated regulation of insulin gene transcription. Mol. Cell. Biol. 19, 8492–8504 10.1128/MCB.19.12.8492 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Liu A., Desai B. M., and Stoffers D. A. (2004) Identification of PCIF1, a POZ domain protein that inhibits PDX-1 (MODY4) transcriptional activity. Mol. Cell. Biol. 24, 4372–4383 10.1128/MCB.24.10.4372-4383.2004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Wong C. K., Wade-Vallance A. K., Luciani D. S., Brindle P. K., Lynn F. C., and Gibson W. T. (2018) The p300 and CBP transcriptional coactivators are required for beta-cell and α-cell proliferation. Diabetes 67, 412–422 10.2337/db17-0237 [DOI] [PubMed] [Google Scholar]
- 42. Golson M. L., and Kaestner K. H. (2017) Epigenetics in formation, function, and failure of the endocrine pancreas. Mol. Metab. 6, 1066–1076 10.1016/j.molmet.2017.05.015 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Xie R., Carrano A. C., and Sander M. (2015) A systems view of epigenetic networks regulating pancreas development and beta-cell function. Wiley Interdiscip. Rev. Syst. Biol. Med. 7, 1–11 10.1002/wsbm.1287 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Bramswig N. C., Everett L. J., Schug J., Dorrell C., Liu C., Luo Y., Streeter P. R., Naji A., Grompe M., and Kaestner K. H. (2013) Epigenomic plasticity enables human pancreatic α to β cell reprogramming. J. Clin. Invest. 123, 1275–1284 10.1172/JCI66514 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Zhang H., Wang W. P., Guo T., Yang J. C., Chen P., Ma K. T., Guan Y. F., and Zhou C. Y. (2009) The LIM-homeodomain protein ISL1 activates insulin gene promoter directly through synergy with BETA2. J. Mol. Biol. 392, 566–577 10.1016/j.jmb.2009.07.036 [DOI] [PubMed] [Google Scholar]
- 46. Eeckhoute J., Briche I., Kurowska M., Formstecher P., and Laine B. (2006) Hepatocyte nuclear factor 4 α ligand binding and F domains mediate interaction and transcriptional synergy with the pancreatic islet LIM HD transcription factor Isl1. J. Mol. Biol. 364, 567–581 10.1016/j.jmb.2006.07.096 [DOI] [PubMed] [Google Scholar]
- 47. Yan C., Yu C., Zhang D., Cui Y., Zhou J., and Cui S. (2016) Protein inhibitor of activated STAT Y (PIASy) regulates insulin secretion by interacting with LIM homeodomain transcription factor Isl1. Sci. Rep. 6, 39308 10.1038/srep39308 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Kim J., Hake S. B., and Roeder R. G. (2005) The human homolog of yeast BRE1 functions as a transcriptional coactivator through direct activator interactions. Mol. Cell 20, 759–770 10.1016/j.molcel.2005.11.012 [DOI] [PubMed] [Google Scholar]
- 49. Shema E., Tirosh I., Aylon Y., Huang J., Ye C., Moskovits N., Raver-Shapira N., Minsky N., Pirngruber J., Tarcic G., Hublarova P., Moyal L., Gana-Weisz M., Shiloh Y., Yarden Y., et al. (2008) The histone H2B-specific ubiquitin ligase RNF20/hBRE1 acts as a putative tumor suppressor through selective regulation of gene expression. Genes Dev. 22, 2664–2676 10.1101/gad.1703008 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50. Batta K., Zhang Z., Yen K., Goffman D. B., and Pugh B. F. (2011) Genome-wide function of H2B ubiquitylation in promoter and genic regions. Genes Dev. 25, 2254–2265 10.1101/gad.177238.111 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51. Fleming A. B., Kao C. F., Hillyer C., Pikaart M., and Osley M. A. (2008) H2B ubiquitylation plays a role in nucleosome dynamics during transcription elongation. Mol. Cell 31, 57–66 10.1016/j.molcel.2008.04.025 [DOI] [PubMed] [Google Scholar]
- 52. Pavri R., Zhu B., Li G., Trojer P., Mandal S., Shilatifard A., and Reinberg D. (2006) Histone H2B monoubiquitination functions cooperatively with FACT to regulate elongation by RNA polymerase II. Cell 125, 703–717 10.1016/j.cell.2006.04.029 [DOI] [PubMed] [Google Scholar]
- 53. Briggs S. D., Xiao T., Sun Z. W., Caldwell J. A., Shabanowitz J., Hunt D. F., Allis C. D., and Strahl B. D. (2002) Gene silencing: trans-histone regulatory pathway in chromatin. Nature 418, 498 10.1038/nature00970 [DOI] [PubMed] [Google Scholar]
- 54. Ng H. H., Xu R. M., Zhang Y., and Struhl K. (2002) Ubiquitination of histone H2B by Rad6 is required for efficient Dot1-mediated methylation of histone H3 lysine 79. J. Biol. Chem. 277, 34655–34657 10.1074/jbc.C200433200 [DOI] [PubMed] [Google Scholar]
- 55. Sun Z. W., and Allis C. D. (2002) Ubiquitination of histone H2B regulates H3 methylation and gene silencing in yeast. Nature 418, 104–108 10.1038/nature00883 [DOI] [PubMed] [Google Scholar]
- 56. Güngör C., Taniguchi-Ishigaki N., Ma H., Drung A., Tursun B., Ostendorff H. P., Bossenz M., Becker C. G., Becker T., and Bach I. (2007) Proteasomal selection of multiprotein complexes recruited by LIM homeodomain transcription factors. Proc. Natl. Acad. Sci. U.S.A. 104, 15000–15005 10.1073/pnas.0703738104 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57. Pagliuca F. W., and Melton D. A. (2013) How to make a functional beta-cell. Development 140, 2472–2483 10.1242/dev.093187 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58. Xie W., Nagarajan S., Baumgart S. J., Kosinsky R. L., Najafova Z., Kari V., Hennion M., Indenbirken D., Bonn S., Grundhoff A., Wegwitz F., Mansouri A., and Johnsen S. A. (2017) RNF40 regulates gene expression in an epigenetic context-dependent manner. Genome Biol. 18, 32 10.1186/s13059-017-1159-5 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59. Cao J., and Yan Q. (2012) Histone ubiquitination and deubiquitination in transcription, DNA damage response, and cancer. Front. Oncol. 2, 26 10.3389/fonc.2012.00026 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60. Herbert T. P., and Laybutt D. R. (2016) A reevaluation of the role of the unfolded protein response in islet dysfunction: maladaptation or a failure to adapt? Diabetes 65, 1472–1480 10.2337/db15-1633 [DOI] [PubMed] [Google Scholar]
- 61. Liu J., Walp E. R., and May C. L. (2012) Elevation of transcription factor Islet-1 levels in vivo increases beta-cell function but not beta-cell mass. Islets 4, 199–206 10.4161/isl.19982 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62. Hunter C. S., and Stein R. (2013) Characterization of an apparently novel beta-cell line-enriched 80–88-kDa transcriptional activator of the MafA and Pdx1 genes. J. Biol. Chem. 288, 3795–3803 10.1074/jbc.M112.434282 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63. Schreiber E., Matthias P., Müller M. M., and Schaffner W. (1989) Rapid detection of octamer binding proteins with 'mini-extracts', prepared from a small number of cells. Nucleic Acids Res. 17, 6419 10.1093/nar/17.15.6419 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64. Ma C., Kojima K., Xu N., Mobley J., Zhou L., Yang S. T., and Liu X. M. (2015) Comparative proteomics analysis of high n-butanol producing metabolically engineered Clostridium tyrobutyricum. J. Biotechnol. 193, 108–119 10.1016/j.jbiotec.2014.10.036 [DOI] [PubMed] [Google Scholar]
- 65. Keller A., Nesvizhskii A. I., Kolker E., and Aebersold R. (2002) Empirical statistical model to estimate the accuracy of peptide identifications made by MS/MS and database search. Anal. Chem. 74, 5383–5392 10.1021/ac025747h [DOI] [PubMed] [Google Scholar]
- 66. Nesvizhskii A. I., Keller A., Kolker E., and Aebersold R. (2003) A statistical model for identifying proteins by tandem mass spectrometry. Anal. Chem. 75, 4646–4658 10.1021/ac0341261 [DOI] [PubMed] [Google Scholar]
- 67. Weatherly D. B., Atwood J. A. 3rd, Minning T. A., Cavola C., Tarleton R. L., and Orlando R. (2005) A heuristic method for assigning a false-discovery rate for protein identifications from Mascot database search results. Mol. Cell. Proteomics 4, 762–772 10.1074/mcp.M400215-MCP200 [DOI] [PubMed] [Google Scholar]
- 68. Liu H., Sadygov R. G., and Yates J. R. 3rd. (2004) A model for random sampling and estimation of relative protein abundance in shotgun proteomics. Anal. Chem. 76, 4193–4201 10.1021/ac0498563 [DOI] [PubMed] [Google Scholar]
- 69. Old W. M., Meyer-Arendt K., Aveline-Wolf L., Pierce K. G., Mendoza A., Sevinsky J. R., Resing K. A., and Ahn N. G. (2005) Comparison of label-free methods for quantifying human proteins by shotgun proteomics. Mol. Cell. Proteomics 4, 1487–1502 10.1074/mcp.M500084-MCP200 [DOI] [PubMed] [Google Scholar]
- 70. Beissbarth T., Hyde L., Smyth G. K., Job C., Boon W. M., Tan S. S., Scott H. S., and Speed T. P. (2004) Statistical modeling of sequencing errors in SAGE libraries. Bioinformatics 20, Suppl. 1, i31–39 10.1093/bioinformatics/bth924 [DOI] [PubMed] [Google Scholar]
- 71. Xu G., Chen J., Jing G., and Shalev A. (2013) Thioredoxin-interacting protein regulates insulin transcription through microRNA-204. Nat. Med. 19, 1141–1146 10.1038/nm.3287 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72. Gupta R., Nguyen D. C., Schaid M. D., Lei X., Balamurugan A. N., Wong G. W., Kim J. A., Koltes J. E., Kimple M. E., and Bhatnagar S. (2018) Complement 1q like-3 protein inhibits insulin secretion from pancreatic beta-cells via the cell adhesion G protein-coupled receptor BAI3. J. Biol. Chem. 293, 18086–18098 10.1074/jbc.RA118.005403 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.